INTRODUCTION
Antimicrobial resistance is a serious threat to the treatment of infectious diseases and is considered one of the leading public health concerns of the 21st century [Reference Webber and Piddock1]. Antibiotic resistance threatens not only the current management of bacterial infections but also the long-term value of antimicrobial agents. The continuing emergence of antimicrobial-resistant Gram-negative pathogens in particular has not been matched by the development of new classes of antimicrobial agents [Reference Patterson2]. Although selective pressure from antimicrobial use is a central factor in the emergence of resistance, transmission of resistant bacteria and/or resistance genes is an important contributor to the prevalence of resistance [Reference vonBaum and Marre3]. Human-to-human transmission as a contributor to the risk of resistance is supported by studies reporting risk factors such as being hospitalized, attending a daycare centre, or living with another person who is colonized with resistant bacteria [Reference Bruinsma4, Reference Reves5]. Transmission via the ingestion of contaminated food or water has also been hypothesized [Reference Prats6, Reference Sørum and L'Abée-Lund7].
Antimicrobial-resistant E. coli have been detected in a variety of food sources including vegetables, meat, and poultry [Reference Johnson8–10] as well as in drinking water [Reference Anderson and Sobsey11, Reference El-Zanfaly12]. However, to date, studies show that vegetarians are as likely to carry resistant strains of E. coli as omnivores [Reference Sannes13, Reference van den Braak14] and no association between the consumption of water contaminated with resistant bacteria and its carriage have been found [Reference Amyes15, Reference Seidman16]. The goal of this study was to assess whether exposure to drinking water contaminated with antimicrobial-resistant E. coli is associated with the carriage of resistant E. coli in the human gastrointestinal tract.
MATERIALS AND METHODS
This study used existing public health water testing infrastructure programmes in the provinces of Ontario and Alberta, Canada, to which users of private water sources submit samples for detection of contamination with E. coli and coliforms. For this study, all E. coli-positive water samples submitted for testing between 1 May 2005 and 30 September 2006 to two regional laboratories in Ontario (London, Hamilton) and Alberta (Calgary, David Thompson) as well as a randomly selected monthly quota of samples from five other Ontario laboratories (Ottawa, Kingston, Peterborough, Orillia, Toronto) were screened for susceptibility to antimicrobial agents.
Subject inclusion
This study drew upon a convenience sample of people living in households that participated in a case-control study to determine the risk factors for contamination of water sources with resistant E. coli. Case households were those with a drinking water sample that tested positive for E. coli resistant to one or more of the antibiotic agents. ‘A’ controls were randomly selected households with water samples yielding E. coli susceptible to antibiotics in the screening panel, and ‘B’ control households were randomly selected from water submissions that were not contaminated with either E. coli or coliforms. Controls were frequency-matched by laboratory region and from samples submitted within 1 month of the date of the case submission. Inclusion was limited to unique households in which at least one adult (aged ⩾18 years) resided on the property from which the water was submitted, spoke English, provided an operational telephone number with the water sample, and consented to share their contact information with the study. Households selected as ‘B’ controls were not eligible if they had a water sample that tested positive for bacterial contamination any time within the previous 12 months.
For the cross-sectional study upon which the following analyses are conducted, all household members who were aged ⩾12 years at the time of the interview and who spent >50% of nights at the household were eligible to participate.
Sample collection
In the London and Hamilton regions, 35% of households were recruited via site visits in which consenting submitters of the water sample completed a household questionnaire. For this cross-sectional study, all consenting household members who provided written consent and agreed to provide a rectal swab were interviewed and asked to provide their swab during the site visit. In the other study regions, and for the remaining 65% of households in the London and Hamilton regions, subjects were interviewed by telephone using a computer-assisted telephone interview system. Rectal swab sampling kits were mailed to all household members who completed a personal questionnaire. Participants mailed swabs to the study laboratory.
This study was reviewed and approved by the institutional review boards of the universities of Western Ontario, Toronto, and Calgary.
Isolation and detection of E. coli
Water samples were tested using membrane filtration and differential coliform agar in Ontario [17] or by the defined substrate method (Colilert®, IDEXX Laboratories, USA) in Alberta. Isolates of E. coli were obtained from defined substrate media by taking a 10 μl aliquot of the presumptive E. coli-positive Colilert sample and swabbing colonies to isolation on X-Gluc agar (Dalynn, Canada) and incubating the samples overnight at 35°C. Up to five presumptive E. coli colonies grown on either differential coliform or X-Gluc agar were pooled, transferred to trypticase soy agar (TSA) slants, and transported to the study laboratory.
Rectal swabs were transported to the study laboratory in Cary–Blair transport media, inoculated into trypticase soy broth (TSB), incubated overnight at 37°C, and archived in skimmed milk at −70°C. Ice crystals from archived TSB suspension were used to inoculate fresh TSB.
Screening assay for drug resistance
Screening for antibiotic resistance was performed by the agar screen plate method. E. coli were cultured in a TSB suspension at 35°C for 4 h before plating 10 μl bacterial culture on MacConkey plates (one control and seven supplemented with antibiotics: gentamicin 8 μg/ml, streptomycin 32 μg/ml, ampicillin 8 μg/ml, nalidixic acid 4 μg/ml, sulfamethoxazole 128 μg/ml, cephalothin 32 μg/ml, tetracycline 4 μg/ml). One resistant lactose-fermenting isolate, preferably from a tetracycline plate (see below), from each water sample was sent to the Laboratory for Foodborne Zoonoses (St Hyacinthe, Canada), where API® test strips (bioMérieux, Canada) were used for confirmation of E. coli. If the isolate was confirmed as E. coli (⩾90% agreement), broth microdilution antimicrobial susceptibility testing was performed and interpreted according to CLSI standards as outlined in the 2004 Canadian Integrated Program for Antimicrobial Resistance report [18], using the National Antimicrobial Resistance Monitoring System (NARMS) Sensititre™ test panel. Antibiotics, with minimum inhibitory concentration defined as resistant, were: ampicillin 32 μg/ml, amoxicillin/clavulanic acid 32/16 μg/ml, cefoxitin 32 μg/ml, ceftiofur 8 μg/ml, ceftriaxone 64 μg/ml, ciprofloxacin 4 μg/ml, nalidixic acid 32 μg/ml, trimethoprim/sulfamethoxazole 4/76 μg/ml, sulfisoxazole 256 μg/ml, tetracycline 16 μg/ml, chloramphenicol 32 μg/ml, amikacin 64 μg/ml, gentamicin 16 μg/ml, kanamycin 64 μg/ml, and streptomycin 64 μg/ml [19].
For rectal swabs, the suspension was incubated for 4 h at 37°C before plating 10 μl onto MacConkey agar plates with and without antibiotics (as described for water testing). One resistant lactose-fermenting isolate, preferably from an ampicillin plate (see below), from each sample was sent to the Laboratory for Foodborne Zoonoses where confirmation of identity and antibiotic testing was conducted as described above for the water samples.
Preliminary studies revealed that >90% of resistant lactose-fermenting isolates were E. coli and that >85% of resistant isolates from the same sample were identical by antibiogram and pulsed-field gel electrophoresis profile regardless of the particular antibiotic plate from which they were cultured. Therefore, a single isolate for testing was selected from the screening plate with the highest frequency of resistance detected: tetracycline for water and ampicillin for rectal swabs.
Data analyses
The prevalence of human carriage of antimicrobial-resistant E. coli was calculated using the total number of specimens yielding E. coli that were resistant to at least one antibiotic divided by the total number of specimens yielding E. coli. Multidrug resistance was defined as resistance to two or more classes of antibiotics. Classes included: β-lactam, tetracycline, sulphonamide, aminoglycoside, quinolone, and chloramphenicol.
For primary multivariable analysis, the dependent variable was whether the E. coli isolated from the rectal swab was resistant to one or more antibiotics in the NARMS panel (defined as antibiotic resistant) vs. being susceptible to all antibiotics tested. The independent variable of primary interest was the consumption of water that was contaminated with antibiotic-resistant E. coli. For this analysis, households that treated their water for bacterial contamination (boiled or treated with chlorine, ultraviolet light, or ozone prior to consumption and for at least 1 year before the sample was taken [20]) were categorized as using uncontaminated water.
It was estimated that, if 1/3 unexposed and 1/2 exposed participants carried an antibiotic-resistant strain of E. coli, 86 people from independent case households and 258 people from independent control households (one case, three controls) were necessary to reject the null hypothesis of ‘no association’ with a power of 80%, given a type I error rate of 5%.
Poisson regression was used since relative risk is the preferred measure for cross-sectional studies and can adequately represent the association between the independent and dependent variables [Reference Lumley, Kronmal and Ma21]. Huber–White robust estimates of variance were used to account for the non-independence of observations within households. Variables associated with the dependent variable at a P value of ⩽0·25 and those with biologically plausible associations and effect modification potential (e.g. antibiotic use, hospitalization, use of bottled water) were used to build the regression model. To maintain precision, continuous variables were kept in original form after confirming that the assumption of linearity of effect in the adjusted regression models was satisfied through examination of residual by predictor plots and failure of the use of a categorical version (as quartiles) to indicate other than a linear association. Regression diagnostics were performed, which included evaluation of dispersion.
RESULTS
A rectal swab was returned by 975/1321 (74%) people who completed a personal questionnaire. Susceptibility testing was completed on 958 swabs (17 swabs were inadequately labelled, lost, or damaged in transport). Study analyses were based on the results of 878 swab-questionnaire pairs since E. coli was not isolated from 46 rectal swabs and 34 swabs could not be matched to all three of the other data points: water test result, household questionnaire, and personal questionnaire.
The 878 participants from 595 households included 431 males and 447 females (age range 12–87 years), 715 of whom lived in Ontario and 163 in Alberta (Table 1). In total, 101 individuals stated they had used an antibiotic in the previous 3 months and 50% of participants had travelled internationally in the previous year, with the USA being the most common destination. Fifty-nine percent (519/878) of participants used water from a source that was contaminated with E. coli at the time of the sampling. Of these, 180 individuals used water contaminated with antimicrobial-resistant E. coli including 94 subjects whose water was untreated, 35 whose water had been treated to kill bacteria for <12 months, and 51 whose water had been treated for >12 months. Overall, 129 individuals used water contaminated with E. coli resistant to one or more antibiotics: 112 (87%) resistant to tetracycline, 69 sulphonamide, 44 β-lactam, 44 aminoglycoside, 13 chloramphenicol, and five resistant to a fluoroquinolone antibiotic. Sixty per cent (78/129) of these participants used water contaminated with strains resistant to two or more classes of antibiotics.
Table 1. Characteristics of individuals with and without antibiotic-resistant E. coli present in rectal swab specimen, bivariate association adjusted for household clustering, Ontario and Alberta, 2005–2006
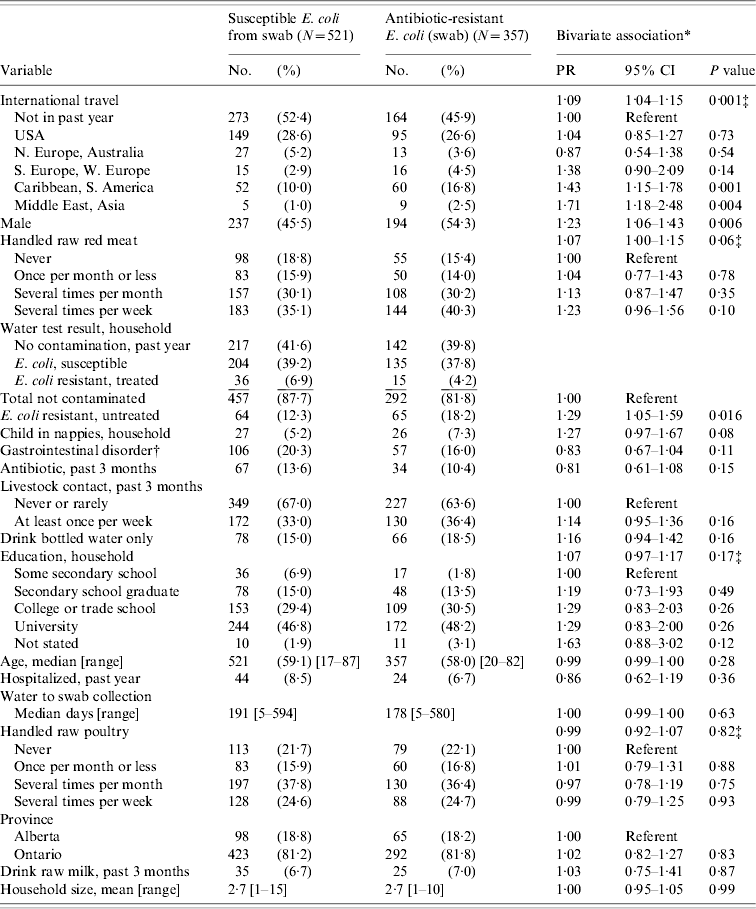
PR, Prevalence ratio; CI, confidence interval.
* Robust variance estimate: standard error adjusted for household clustering.
† Gastrointestinal disorders: Crohn's disease, celiac disease, irritable bowel syndrome, colitis, ileitis, ulcers, gastroesophageal reflux.
‡ Probability that at least one of the variables' regression coefficients≠0.
Of the rectal swabs with E. coli isolated, 357/878 (41%) were resistant to one or more of the study antibiotics (Table 2). Resistance was highest to β-lactam, tetracycline, sulphonamide, and aminoglycoside agents with lower rates of resistance to fluoroquinolones and chloramphenicol. The rates of resistance were similar for subjects in Ontario (292/715, 41%) and Alberta (65/163, 40%). There was no statistically significant difference in rates of resistance for participants using water that had been free of bacterial contamination for ⩾1 year (142/359, 40%), those whose water tested positive for E. coli susceptible to all antibiotics in the screening panel (135/339, 40%), and those whose water was contaminated with antimicrobial-resistant E. coli but who had been treating that water for ⩾1 year prior to the rectal sample (15/51, 29%). These 749 people, who used water not contaminated with resistant E. coli, were combined for analyses comparing them with 129 people consuming water contaminated with resistant E. coli at, or within 1 year of, rectal swab collection.
Table 2. Results of laboratory testing of rectal swabs, by water test result, unadjusted, Ontario and Alberta, 2005–2006
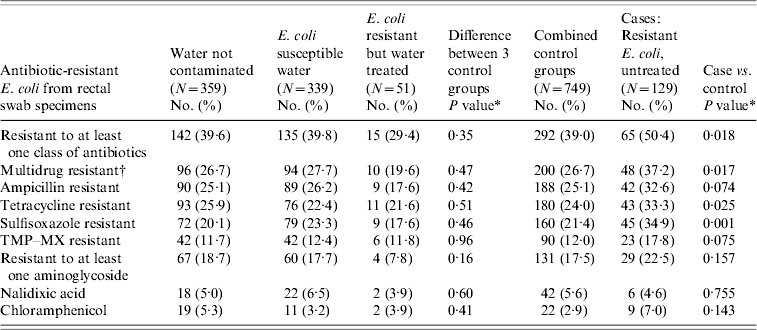
TMP–SMX, Trimethoprim–sulfamethoxazole.
* P value based on Pearson's χ2, unadjusted for household clustering.
† Resistant to two or more classes of antibiotics.
Of the 230 people carrying E. coli resistant to a β-lactam, 18/44 (41%) were exposed to water contaminated with E. coli resistant to a β-lactam compared to 212/834 (25%) who were not. This association was statistically significant after adjusting for household clustering, with a prevalence ratio (PR) of 1·6 [95% confidence interval (CI) 1·1–2·4, P=0·02]. A statistically significant association was also found for tetracycline (PR 1·6, 95% CI 1·2–2·1, P=0·002), sulphonamides (PR 1·9, 95% CI 1·3–2·6, P=0·001), and chloramphenicol (PR 5·2, 95% CI 2·1–13·3, P=0·001). No association was found for aminoglycosides (P=0·21) or fluoroquinolones (PR 3·7, 95% CI 0·8–16·6, P=0·09). Of note, only five people carrying fluoroquinolone-resistant E. coli were exposed to untreated water sources.
Of the 101 participants who used an antibiotic in the 3 months prior to rectal swab collection, 85 were able to name the antibiotic(s) they used including β-lactams (n=42), fluoroquinolones (n=9), tetracyclines (n=7), macrolides (n=6), and aminoglycoside (n=1) while four participants named two antibiotics and one named three. The use of a fluoroquinolone within 3 months of collection of the rectal swab was associated with carriage of fluoroquinolone-resistant E. coli (PR 3·8, 95% CI 1·1–13·5, P=0·04). No association was found between the use of β-lactam agents and the carriage of β-lactam-resistant E. coli (P=0·55), tetracycline and the carriage of tetracycline-resistant E. coli (P=0·98), or between macrolide agents and the carriage of any specific resistances.
Bivariate analyses showed a statistically significant association between the carriage of an antibiotic-resistant strain of E. coli and living in a dwelling served by a water source contaminated with antibiotic-resistant E. coli that had not been treated to kill bacteria for at least 12 months (Table 1). Carriage was also associated with the sex of the respondent, with males having higher rates of carriage than females. Participants who travelled internationally within 1 year of swab collection were also more likely to carry a resistant strain of E. coli.
While holding the effects of other variables in the multivariable Poisson regression model constant, people living in households that used untreated antimicrobial-resistant E. coli-contaminated water were 26% more likely to be colonized with antimicrobial-resistant E. coli than people living in households with water sources that were not contaminated or that were contaminated but were treated for ⩾12 months (Table 3). Other variables in the model include sex (with males at higher risk than females), frequent handling of raw red meats (beef, pork, and/or lamb), having a child in nappies living in the household, and international travel in the previous year. Travel to the Middle East and South East Asia, including China and Japan, was associated with the highest prevalence ratio.
Table 3. Multivariable model of association between faecal carriage of antimicrobial-resistant E. coli and covariates; Poisson regression adjusted for household clustering, Alberta and Ontario, 2005–2006
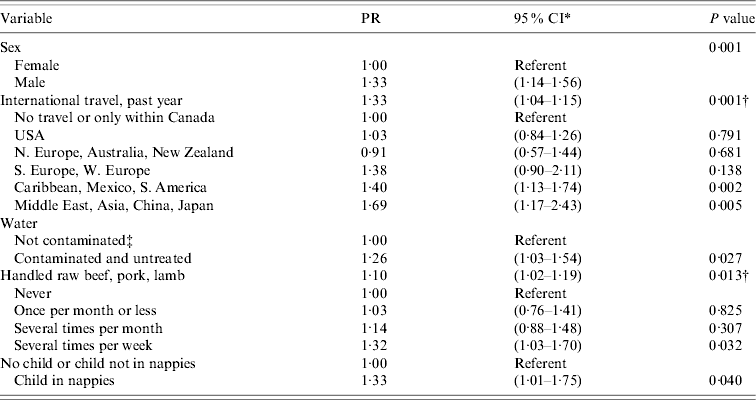
PR, Prevalence ratio; CI, confidence interval.
* Robust variance estimate with standard error adjusted for household clustering.
† Probability that at least one of the variables' regression coefficients≠0.
‡ Water source that was uncontaminated (no bacteria) for ⩾1 year, or water tested positive for E. coli that was sensitive to screening panel of antibiotics, or water contaminated with antibiotic-resistant E. coli and treated (boiled, chlorinated, UV, ozone, or candle/ceramic) for ⩾1 year.
DISCUSSION
This study found that the prevalence of carriage of antimicrobial-resistant E. coli was 26% higher in study participants who consumed water contaminated with antimicrobial-resistant E. coli than in those using uncontaminated or adequately treated water, after adjusting for the impact of other associated risk factors. Two previous studies, both conducted in India, investigated the contribution made by contaminated water to the high rates of faecal carriage of antimicrobial-resistant bacteria in various villages [Reference Amyes15, Reference Seidman16]. However, comparisons of carriage of resistant bacteria were not feasible because, in both studies, almost all of the bacteria identified in communal water sources were antimicrobial resistant.
In North America, about 12% of the population – over 4 million Canadians and 33 million Americans [Reference Corkal, Schutzman and Hilliard22, 23] – rely on private drinking water sources, as do hundreds of millions of people across the globe, many of whom live in countries without the resources for adequate water treatment. In some studies, the rates of antimicrobial resistance in E. coli in drinking water supplies have been reported to be as high as 36% [Reference Amyes15, Reference Seidman16, Reference Walia24]. The risk for transmission exists even in usually safe water sources if unusual circumstances such as heavy rainfall and/or failure to adequately treat water occur, as seen during the Walkerton, Ontario outbreak in 2000 where seven people died of E. coli O157:H7 and in Milwaukee, USA in 1993 where 54 people of died of cryptosporidiosis [Reference Auld, MacIver and Klaassen25].
Our study focused on the single bacterial species (E. coli) that is used to assess the likelihood of faecal contamination of water supplies. Antibiotic resistance genes have been shown to be transferable across different strains of E. coli and between E. coli and other Enterobacteriaceae [Reference Walia24]. However, other bacteria are more common causes of drinking water contamination than E. coli such that exposure to, and subsequent carriage of, resistant bacteria from contaminated water sources may be considerably more prevalent than is implied in this study. The list of strategies to reduce the prevalence and transmission of antimicrobial resistance needs to include the adequate treatment of contaminated water and appropriate management of all drinking water sources to prevent contamination.
Participants in our study who had travelled outside of North America and northern European countries within 12 months of the swab sample were significantly more likely to carry resistant strains of E. coli than other participants. Several other studies have also found an association between international travel and higher rates of carriage or infection with resistant bacteria, including studies that sampled students before and after travelling [Reference Sannes13, Reference Murray26]. This may be related to high colonization pressure, which increases the probability of person-to-person transmission, or the ingestion of resistance-carrying bacteria via contaminated food and water [Reference Bonten27].
Although some previous studies have, like ours, detected a higher prevalence of carriage in males than females [Reference Leflon-Giubout28], the opposite has also been reported [Reference Hay29], and others report no difference by sex [Reference Hannah30]. In our study, males and females were equally likely to be exposed to other putative risk factors for the carriage of antimicrobial-resistant E. coli, suggesting that there may have been unmeasured factors driving the difference between the sexes.
Participants living with children in nappies at the time of the sampling were significantly more likely to carry a resistant strain of E. coli than other subjects. This is not unexpected: in studies of healthy children, a higher proportion of children aged <2 years had antibiotic-resistant E. coli detected in faecal [Reference Reves5] and urine [Reference Storby, Osterlund and Kahlmeter31] samples than older children and at least two other studies of household clustering of resistant E. coli noted that strains were often shared between adults and toddlers [Reference Johnson32, Reference Lietzau33].
In our study, participants who handled raw red meat (beef, lamb, pork) several times per week were more likely to carry a resistant strain of E. coli than people who handled it less often. This association may be explained by the observation that 12% of beef and 19% of pork retail meat samples taken in 2006 in Canada were contaminated with antimicrobial-resistant E. coli [10]. However, one would expect similar results for participants who frequently handled poultry products since an even higher proportion of 2006 retail samples of chicken (26%) were contaminated with resistant bacteria [10]. Recent studies have shown similar genomic determinants in E. coli samples from human samples and those from poultry, pork, and beef retail meats [Reference Vincent34, Reference Hannah35], suggesting that either handling or consuming meat and poultry might result in transmission. However, in other studies, vegetarians have been shown to have similar rates of carriage of resistant E. coli as people consuming meat and poultry products [Reference Sannes13, Reference Guinée, Ugueto and van Leeuwen36] and the one study that tried to separate the effects of exposure to poultry vs. other red meats found no difference [Reference Sannes13].
The current or recent use of antibiotics has been identified as a risk factor for human carriage in most, but not all, studies and not for all antimicrobial agents [Reference Sannes13, Reference Hannah30, Reference Akwar37]. Recent exposure is more likely to be associated with resistance to antibiotics where resistance is associated with mutations, or the acquisition of plasmids or transposons, and less likely to be associated with resistance when the acquisition of resistance requires more complex genetic events [Reference Boerlin and Reid-Smith38]. Consistent with other studies, recent use of fluoroquinolones was associated with carriage of fluoroquinolone-resistant E. coli, but recent use of β-lactam antibiotics had no impact on β-lactam resistance [Reference Nijssen39]. However, the carriage of resistant E. coli is transient following the use of antibiotics – with duration of carriage reported lasting from weeks to months [Reference Raum40, Reference Maslow, Lee and Lautenbach41] making it necessary to conduct large prospective cohort studies to adequately determine the association between specific antibiotic use and the subsequent carriage of resistant bacteria.
This study has a number of limitations. In Canada, private water sources are not registered and the owners are not required to test their water for microbial contamination; thus, this study is based on a convenience sample of people living in households from which water was submitted for bacteriological testing. Moreover, although the prevalence of carriage did not appear to vary with age in our sample, the age distribution of respondents may have influenced the prevalence of risk factors for colonization [Reference Hannah30, Reference Degener42]. It is also not possible to tell whether the association between carriage and water contamination would have been stronger if our rectal swab samples had been obtained closer to the time of the water samples or if the association was present because water contamination was persistent. However, no effect of the time lag between submission of water samples and rectal swabs was detected on the association between antibiotic resistance in the two, although our power to assess this was limited. Finally, the method of selecting isolates from screening plates would not have detected all resistant isolates. Thus, the estimates of prevalence of resistance, in both water and rectal swabs, will be underestimates and may underestimate the degree of association between the two.
The prevalence of faecal carriage of antimicrobial-resistant E. coli in 878 non-institutionalized Canadian subjects who used private water sources was 41%, with 26% carrying strains resistant to ampicillin and 5·5% carrying strains resistant to nalidixic acid. The proportion of people carrying resistant strains of E. coli in our study was higher than that reported by two previously published studies of non-institutionalized Canadians. Bruinsma et al. reported that 22% and 1% of participants carried amoxicillin and nalidixic acid-resistant strains of E. coli, respectively [Reference Bruinsma4], while Akwar et al. reported that 16% and 0·2% of participants carried amoxicillin and nalidixic acid resistance, respectively [Reference Akwar37]. Although differing breakpoints and detection methods make direct comparisons problematical, it appears that there may be an increase in the prevalence of carriage of resistant strains of bacteria in non-institutionalized people living in Canada, a nation with relatively strict regulations on the distribution of antimicrobial agents.
It is clear from our findings that resistance to antibiotics of human importance has made its way from the clinical setting to the general population. This is troubling for several reasons. The transmission potential of resistant bacteria and the genes that confer resistance increases exponentially as the prevalence increases in the general population. Moreover, the treatment of infections is hampered since the number of antibiotics available to treat them is reduced and empirical therapy must be done with an eye on the ever-evolving local rates of resistance. Medical practitioners can help reduce the prevalence of resistant bacteria through judicious use of antimicrobial agents. However, that is just one piece of this complex issue. Research must continue into determining the risk factors and preventive mechanisms for the emergence, persistence, and transmission of antimicrobial resistance. According to our results, the arsenal of resistance prevention mechanisms must include the universal treatment of contaminated water sources.
APPENDIX
Antimicrobial Resistant Organisms (ARO) in Water Study Group
D. Daignault, B. Crago, S. Braithwaite, C. Guénette, K. Grimsrud, P. Cantin, M. Jerrett, P. Michel, M. Buzzelli, M. Mulvey, P. Lavallois, F. Ruf, B. Cieben, A. Chagla, R. Irwin.
ACKNOWLEDGEMENTS
This work was support by the Canadian Institutes of Health Research (CIHR-IPPH SWF 66539), the physicians of Ontario through the Physicians' Services Incorporated Foundation (grant no. 0507), and the Alberta Heritage Foundation for Medical Research (grant no. 200400989). We thank the staff at the collaborating public health laboratories including the Alberta ProvLab and the London, Hamilton, Kingston, Orillia, Ottawa, Peterborough, and Toronto public health laboratories in Ontario, as well as the Safe Water Unit at the Ontario Ministry of Health & Long Term Care. We acknowledge the efforts of the research staff and students at the universities of Calgary, British Columbia, Manitoba, Montréal, Toronto, McMaster, and Western Ontario, as well as the support of the staff of the public health units in Ontario and health regions of Alberta.
DECLARATION OF INTEREST
None.





