Introduction
Cacao (Theobroma cacao L.) is cultivated in the humid tropics for its fruits (pods) from which the seeds (beans), following fermentation and drying are used to produce chocolate and other confectionery products. Cacao from the South American Amazon basin as its primary centre of diversity (Cheesman, Reference Cheesman1944; Schultes, Reference Schultes and Stone1984; Bartley, Reference Bartley2005) has been repeatedly introduced into various cacao growing countries across the globe (Schultes, Reference Schultes and Stone1984; Kennedy and Mooleedhar, Reference Kennedy and Mooleedhar1993; Lockwood and End, Reference Lockwood and End1993; Bartley, Reference Bartley2005). Presently, West Africa contributes over 70% to the global cacao production, with Cote d'Ivoire and Ghana being the major producing countries (ICCO, 2021).
Cultivation of cacao in Ghana commenced in the 19th century where the available cultivars were mainly of Amelonado origin and uniform in most economic traits (Posnette, Reference Posnette1943). In 1938, research into cacao formally began in Ghana through the establishment of the West Africa Cacao Research Institute following the destruction of cacao by the cocoa swollen shoot virus disease (CSSVD) and the difficulties associated with the crop's establishment resulting from primary forest cover loss (Posnette, Reference Posnette1941). To broaden the genetic base of the existing West African cacao germplasm composed mainly of Amelonado, different cacao introductions have occurred (Posnette, Reference Posnette1951; Glendinning, Reference Glendinning1957; Lockwood and Gyamfi, Reference Lockwood and Gyamfi1979; Abdul-Karimu et al., Reference Abdul-Karimu, Adomako and Adu-Ampomah2006). Some of these introductions were evaluated for agronomic performance and promising clones recommended as parents for production of hybrids for farmers (Posnette, Reference Posnette1951; Glendinning, Reference Glendinning1964, Reference Glendinning1966; Lockwood, Reference Lockwood1971; Adomako et al., Reference Adomako, Allen and Adu-Ampomah1999, Reference Adomako, Padi, Opoku, Assuah, Domfeh and Owusu-Ansah2007).
Based on the classification of cacao by Motamayor et al. (Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008), the available cacao germplasm (about 1000 clones) in Ghana was separated into eight genetic groups; namely, Nanay, Iquitos, Guiana, Amelonado, Trinitario, Purús, Contamana/Scavina and Marañón (Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015). The authors further observed that the base of the farmers' varieties in Ghana has been limited to few clones from only four genetic groups. Besides the narrow genetic base, the farmers' varieties were released many years ago (Thresh et al., Reference Thresh, Owusu, Boamah and Lockwood1988; Abdul-Karimu et al., Reference Abdul-Karimu, Adomako and Adu-Ampomah2006). The large-scale production of these varieties amidst changing climatic conditions (Sala et al., Reference Sala, Cilas, Gimeno, Wohl, Opoku, Găinuşă-Bogdan and Ribeyre2021) could partly explain the low average yields of 400 kg/ha observed in farms in Ghana (Aneani and Ofori-Frimpong, Reference Aneani and Ofori-Frimpong2013). Therefore, there is need to broaden the genetic base of the farmers' varieties by introducing superior clones of diverse genetic backgrounds in the seed gardens for production of improved varieties with better adaptation and yield. To this end, a recurrent selection programme involving superior clones in the available germplasm (largely from underrepresented genetic groups) was implemented, to identify new clones with good combining abilities for survival, vigour, precocity, yield, and disease resistance (Padi et al., Reference Padi, Ofori and Arthur2017b, Reference Padi, Domfeh, Arthur and Ofori2018). On the basis of the selection theory by Simmonds (Reference Simmonds1996), which suggests that the best clones are more frequently obtained in the best families as confirmed by Padi et al. (Reference Padi, Adu-Gyamfi, Akpertey, Arthur and Ofori2013a, Reference Padi, Takrama, Opoku, Dadzie and Assuah2013b), promising individual trees (ortets) were selected from families showing outstanding performance for a number of agronomic traits. Assessment of genetic diversity would be useful in revealing the genetic relationships among the current selections to guide their effective conservation, management, and utilization in the cacao breeding programme.
In Ghana and other cacao growing countries in West Africa such as Côte d'Ivoire, Nigeria and Cameroon, cacao planting materials are supplied to farmers as seed pods from established seed gardens planted with recommended parental clones. At present, there are about 26 cacao seed garden stations in Ghana being managed by the Seed Production Division (SPD) of the Ghana Cocoa Board. Each seed garden station is generally planted with between four and six recommended parental clones which are manually pollinated to produce seedling varieties of good agronomic performance for farmers. Padi et al. (Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) observed that 2 to 100% of trees in clonal plots sampled across six cacao seed gardens in Ghana were mislabelled. More importantly, studies on cocoa (Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) and coffee (Akpertey et al., Reference Akpertey, Padi, Meinhardt and Zhang2020) revealed that progenies derived from mislabelled or pollen contaminated clones had reduced vigour and number of pods per tree relative to those with correct parentage. For this reason, it is essential to frequently monitor the genetic purity of seedling varieties produced in the seed gardens and implement measures, where necessary, to maintain the genetic integrity of varieties distributed to farmers.
Single nucleotide polymorphism (SNP) markers have become the marker of choice for genetic studies in T. cacao due to their cost-effectiveness, high abundance in the genome and amenability to high-throughput automation (Gupta et al., Reference Gupta, Roy and Prasad2001; Mammadov et al., Reference Mammadov, Aggarwal, Buyyarapu and Kumpatla2012). In recent times, the effectiveness of SNP fingerprinting in improving the efficiency of variety development and recommendations in cacao breeding programmes has been undoubtedly proven in cacao accessions in Honduras and Nicaragua (Ji et al., Reference Ji, Zhang, Motilal, Boccara, Lachenaud and Meinhardt2012), Indonesia (Lukman et al., Reference Lukman, Susilo, Dinarti, Bailey, Mischke and Meinhardt2014), Ghana (Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015), Puerto Rico (Cosme et al., Reference Cosme, Cuevas, Zhang, Oleksyk and Irish2016), Colombia (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017), Costa Rica (Mata-Quirós et al., Reference Mata-Quirós, Arciniegas-Leal, Phillips-Mora, Meinhardt, Motilal, Mischke and Zhang2018), Uganda (Gopaulchan et al., Reference Gopaulchan, Motilal, Bekele, Clause, Ariko, Ejang and Umaharan2019), Dominica (Gopaulchan et al., Reference Gopaulchan, Motilal, Kalloo, Mahabir, Marissa, Joseph and Umaharan2020), tropical Americas (Gutiérrez et al., Reference Gutiérrez, Martinez, Zhang, Livingstone, Turnbull and Motamayor2021), and north Peru (Bustamante et al., Reference Bustamante, Motilal, Calderon, Mahabir and Oliva2022). These authors used varying number of informative SNP markers ranging from 44 to 219 in achieving different objectives, including identification of mislabelled/duplicated accessions, genetic diversity, ancestry, population structure and parentage analyses, and construction of core collections in cacao.
The objectives of this study were to (1) determine the population structure and parentage of locally bred cacao accessions in Ghana and (2) verify the parentage of seedling varieties produced in seed gardens for commercial plantations.
Materials and methods
Cacao sample analysis and SNP genotyping
Two populations, consisting of 168 ramets (pop1) and 752 bi-clonal seedlings (pop2) were sampled and genotyped in the current study. The ramets were developed from ortets selected from progeny trials established on-station and on-farm by the Cocoa Research Institute of Ghana (CRIG) from 2010 to 2021. Ortets were selected from families generated from parents with good combining abilities and showing outstanding performance for a number of agronomic traits including yield, growth, precocity and adaptability to marginal production conditions (Padi et al., Reference Padi, Takrama, Opoku, Dadzie and Assuah2013b, Reference Padi, Ofori and Akpertey2017a; Ofori et al., Reference Ofori, Padi, Akpertey, Asare Bediako, Arthur, Adu-Gyamfi, Nyadanu, Obeng-Bio and Anokye2023). Generally, ortets selection was based on visual inspection for number of pod-harvest scars, and absence of disease symptoms, including stem canker (caused by Phytophthora spp.), and cocoa swollen shoot virus and black pod diseases. The bi-clonal seedlings were generated through manual pollination by the Seed Production Division (SPD) of the Ghana Cocoa Board to supply planting material of recommended varieties for commercial plantations. Additionally, 41 international clones (from the CRIG germplasm) belonging to the 10 genetic groups described by Motamayor et al. (Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008) and Trinitario group were included as references for genetic structure and parentage analyses of local clones (Table 1). Sources of reference accessions and references for the genetic clusters are presented in Table S1.
Table 1. Reference cacao clones of 11 genetic groups used in genetic structure and parentage analyses for assignment of ancestry

Young leaves of tagged trees/seedlings were collected into labelled brown paper envelopes. Total genomic DNA was extracted from the leaf samples using CTAB protocol (Doyle and Doyle, Reference Doyle and Doyle1990). Concentration of DNA was determined with a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Genotyping was done at 45 and 60 SNP loci for the ramets and bi-clonal seedlings, respectively. The SNP markers were selected from 1560 candidate SNPs developed from cDNA sequences from a wide range of cocoa tissues (Argout et al., Reference Argout, Fouet, Wincker, Gramacho, Legavre, Sabau, Risterucci, Da Silva, Cascardo, Allegre, Kuhn, Verica, Courtois, Loor, Babin, Sounigo, Ducamp, Guiltinan, Ruiz, Alemanno, Machado, Phillips, Schnell, Gilmour, Rosenquist, Butler, Maximova and Lanaud2008). Selection of markers was based on their distribution across the ten chromosomes of cacao and levels of polymorphism in the previous experiments (Ji et al., Reference Ji, Zhang, Motilal, Boccara, Lachenaud and Meinhardt2012; Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015; Mata-Quirós et al., Reference Mata-Quirós, Arciniegas-Leal, Phillips-Mora, Meinhardt, Motilal, Mischke and Zhang2018). The discovery, design, and map positions of these SNP markers on the T. cacao consensus linkage map are given in Allegre et al. (Reference Allegre, Argout, Boccara, Fouet, Roguet, Bérard, Thévenin, Chauveau, Rivallan, Clement, Courtois, Gramacho, Boland-Augé, Tahi, Umaharan, Brunel and Lanaud2012). The list of the SNPs and their flanking sequences are presented in Table S2. Leaf samples (as desiccated leaf discs) from which DNA was extracted for SNP assay were obtained from each cacao accession in 96-well plates that were supplied by KBiosciences, UK. The subsequent SNP fingerprinting was performed at KBiosciences using the competitive allele-specific PCR KASPar chemistry (KBiosciences, Hoddesdon, Hertfordshire, UK).
Data analyses
Descriptive statistics for measuring informativeness of the SNP markers, including observed heterozygosity, expected heterozygosity, polymorphic information content (PIC) and the probability of identity among siblings (PID-sib) were estimated using CERVUS v3.0.7 (Kalinowski et al., Reference Kalinowski, Taper and Marshall2007). The PID-sib is defined as the probability that two sibling individuals drawn at random from a population have the same multilocus genotype (Waits et al., Reference Waits, Luikart and Taberlet2001). In addition, major allele frequency was computed for the SNP markers using PowerMarker v3.25 (Liu and Muse, Reference Liu and Muse2005). For the identification of duplicates, pairwise multilocus matching was performed among individual samples in CERVUS v3.0.7 using the Identify Analysis module. Clones with different names that were fully matched at all SNP loci were declared duplicates.
Population structure of the 168 cacao clones was determined using the Bayesian model-based clustering algorithm of STRUCTURE v2.3.4 (Pritchard et al., Reference Pritchard, Stephens and Donnelly2000). SNP fingerprints at the 45 loci of 22 pure reference accessions, each composed by two clones representing the 10 genetic groups identified by Motamayor et al. (Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008) and Trinitario, were included in the analysis to trace the possible ancestry in the local clones. An admixture model with alpha inferred, independent allele frequency with 200,000 burn-ins and 500,000 Markov Chain Monte Carlo (MCMC) repetitions was used. The number of clusters (K) was set from 4 to 10 and ten independent runs were performed for each value of K. The optimum K value was determined using the ad hoc ΔK method described by Evanno et al. (Reference Evanno, Regnaut and Goudet2005) as implemented in the STRUCTURE HARVESTER software (Earl and von Holdt, Reference Earl and von Holdt2012).
Further clustering of the 168 local cacao clones in relation to the 22 pure reference accessions as used for the STRUCTURE was performed using principal coordinates analysis (PCoA) in GenAlEx v6.5 (Peakall and Smouse, Reference Peakall and Smouse2006, Reference Peakall and Smouse2012). Pairwise Euclidean distances (GD) were calculated on the 45 SNP data and the GD was used to perform the PCoA using a covariance matrix with data standardization option.
Parentage analysis was conducted to assess the parentage contribution from the reference clones to the local clones and identify parent-offspring trios for the bi-clonal seedlings (with unknown maternity and paternity). A likelihood-based method implemented in CERVUS v3.0.7 (Marshall et al., Reference Marshall, Slate, Kruuk and Pemberton1998; Kalinowski et al., Reference Kalinowski, Taper and Marshall2007) was used for computation. The list of candidate parents for the clones included 33 pure reference accessions each composed by three clones belonging to the 10 genetic groups (Motamayor et al., Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008) and Trinitario. For the bi-clonal seedlings, the list of candidate parents comprised all genotypes found in trees of the core clone selection which had been used as progenitors in seed gardens or breeding programme in Ghana. In CERVUS, an error rate of 0.01 was used as the proportion of mistyped loci. In the parentage analysis, simulations were run for 20,000 cycles with the assumption that 90% of the candidate parents were sampled (80% was assumed for the local clones), and a total of 95% of loci were typed. Critical likelihood values (LOD scores) of 95% (strict) and 80% (relaxed) confidence in assignments were obtained using simulations.
Results
Genetic diversity statistics
Two (TcSNP 1075 and TcSNP 1096) of the 60 SNP markers used to genotype the bi-clonal seedlings were found to be monomorphic and were eliminated from further analysis. The PIC, major allele frequency, observed heterozygosity and expected heterozygosity of the SNP markers employed in the analysis of the local clones, reference clones and bi-clonal seedlings are presented in Tables S3–S5. The clones and bi-clonal seedlings showed mean PIC of 0.277 ± 0.016 and 0.256 ± 0.017, respectively. Mean major allele frequency of the clones was 0.737 ± 0.021, whereas the bi-clonal seedlings had 0.783 ± 0.022. Average observed heterozygosity values of 0.378 ± 0.026 and 0.348 ± 0.025 were obtained respectively for clones and bi-clonal seedlings. Mean expected heterozygosity was 0.349 ± 0.022 for the clones and 0.323 ± 0.022 for the bi-clonal seedlings. The local clones had higher mean observed heterozygosity (0.378 ± 0.026) than the reference population (0.187 ± 0.018), indicating that the ramets were derived from hybrid families (ortets). The probability of identity among siblings (PID-sib) based on the 45 and 58 SNP loci were 4.43 × 10−8 and 1.15 × 10−9, respectively, indicating that there is almost a null probability of finding two individuals with the same genotype in the populations. Additionally, a high total exclusion probability of 99.999 × 10−2 was obtained for the 45 and 58 SNP markers used for parentage analysis of clones and seedling varieties, respectively. This indicates that the set of markers are sufficient for assignment of parentage in cacao. The 168 local selections were all distinct clones without duplication.
Inference of ancestral background of local clones
The Bayesian clustering analysis produced five clusters (K = 5), the optimal value determined by the ad hoc ΔK statistic (Evanno et al., Reference Evanno, Regnaut and Goudet2005) (Fig. S1). These clusters are associated with clones of the 10 genetic groups (Motamayor et al., Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008) and Trinitario used as controls in this study. At the threshold of Q = 0.80 (Fig. 1, Table S6), cluster 1 (red) comprised reference accessions from Guiana and Marañón with 35 (20.8%) local selections. Cluster 2 (green) was composed of reference samples from Iquitos with 18 (10.7%) local clones. Cluster 3 (blue) grouped reference accessions from Nacional, Criollo and Curaray genetic backgrounds with no local sample. Cluster 4 (yellow) consisted of reference accessions from Contamana and Purús with 48 (28.6%) local clones. The fifth cluster (pink) included reference samples from Amelonado, Trinitario and Nanay genetic backgrounds, and 21 (12.5%) local accessions. At Q < 0.80, 16 (9.5%), 11 (6.5%), 5 (3.0%) and 14 (8.3%) samples of admixed parentage were found in clusters 1, 2, 4 and 5, respectively. In general, ancestral contributions to the local clones were in the decreasing order; Contamana-Purús > Guiana-Marañón > Amelonado-Trinitario-Nanay > Iquitos (Fig. 1, Table S6).
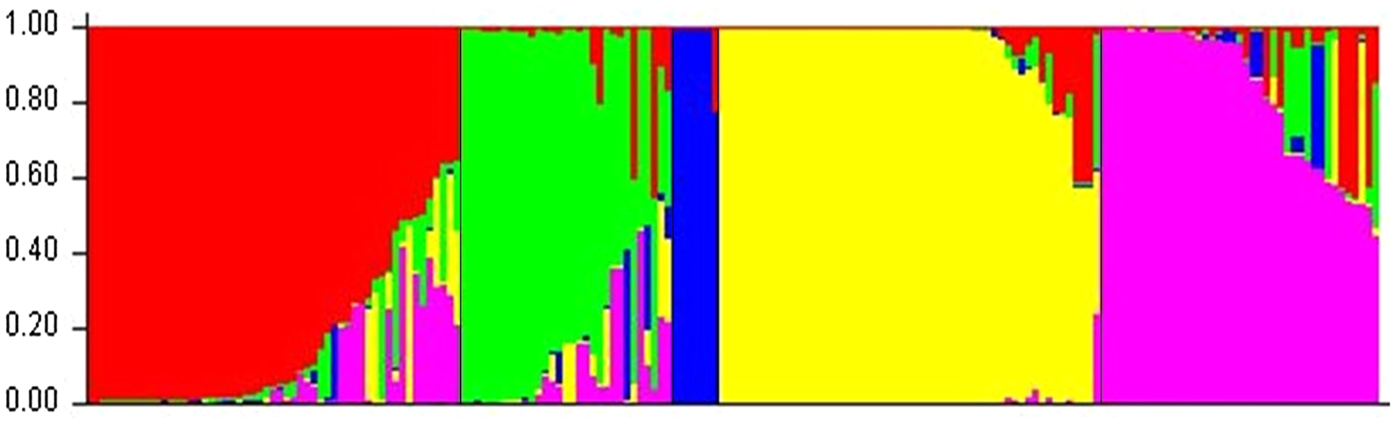
Figure 1. Q-plot showing clustering of 168 local and 22 reference T. cacao accessions based on analysis of genotypic data at 45 SNP loci using STRUCTURE (K = 5). Each genotype is represented by a vertical bar. The coloured subsections within each vertical bar indicate membership coefficient (Q) of the genotype to different subpopulations. Identified subpopulations following Motamayor et al. (2008) are Guiana-Marañón (red): GU 216/A, GU 230/A, PA 150, PA 70; Iquitos (green): IMC 33, IMC 6; Nacional-Criollo-Curaray (blue): Gloria 1, Gloria 12, PR145, PR71, LCT EEN 389, LCT EEN 434; Contamana-Purús (yellow): Ucayali 66, Ucayali 69, LCT EEN 411, LCT EEN 414; Amelonado-Trinitario-Nanay (pink): ES 03 Belize, PR76, ICS 40, RIM 189, POUND 1/B, POUND 2/A. At Q < 0.80, admixed samples in subpopulations 1, 2, 4, and 5 were 16, 11, 5, and 14, respectively.
Parentage analysis shows that 22 known parental clones were responsible for the maternity or paternity of 152 local clones (90% of 168 accessions) at the confidence level above 80%. When the confidence level was raised to 95%, the number of identified parent-offspring relationships was reduced to 113 with contributions from 21 parental clones (Table S7). Among the 152 identified parent-offspring relationships, 43 (28.3%) were associated with Contamana parents, 40 (26.3%) with Marañón, 26 (17.1%) with Guiana, 15 (9.9%) with Amelonado, 15 (9.9%) with Iquitos, 8 (5.3%) with Trinitario, 3 (2.0%) with Nanay and 2 (1.3%) with Purús (Fig. 2, Table S7). The assigned parent-offspring relationships largely agree with the results of the genetic structure (Fig. 1), reaffirming the ancestral contributions of Guiana, Marañón, Contamana, Purús, Amelonado, Trinitario, Nanay and Iquitos genetic groups to the parentage of the local clones. However, with the disintegration of the joint genetic clusters by the likelihood-based parentage analysis, the contributions of the eight genetic groups were in the decreasing order; Contamana > Marañón > Guiana > Amelonado and Iquitos > Trinitario > Nanay > Purús (Fig. 2).
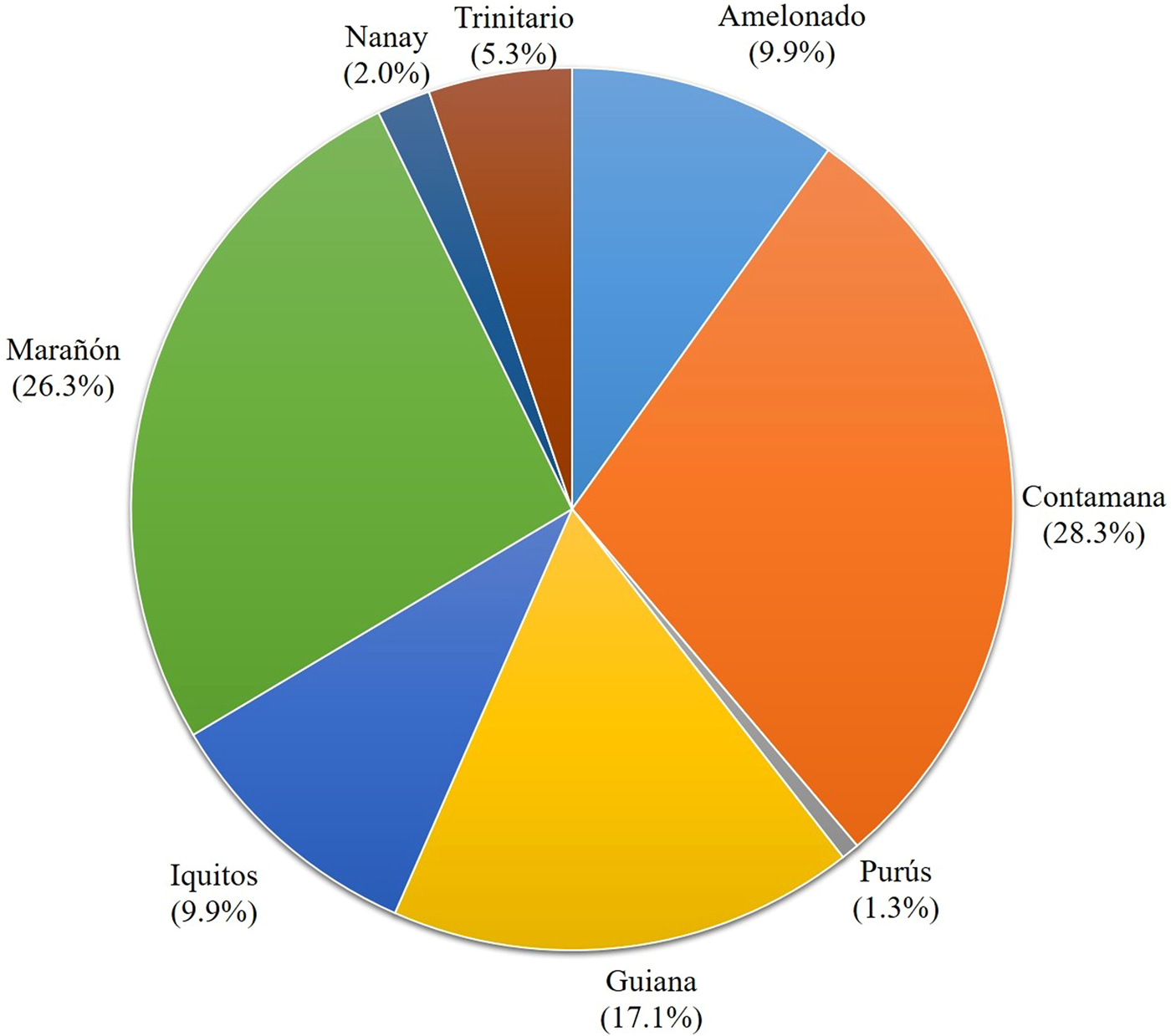
Figure 2. Partitioned ancestry based on assigned parentage of 152 cacao accessions from Ghana with LOD scores above 80% probability. Critical LOD (the natural logarithm of the likelihood) ratios for assignment of maternity/paternity are 4.17 at >95% confidence and 0.33 at >80% confidence.
Genetic relationship between the local selections and the reference clones
The principal coordinates analysis (PCoA) showed the genetic relationships between the local selections and reference clones (Fig. 3). The first and second axes explained 25.7 and 9.8% of the total variation, respectively. Consistent with the results of the Bayesian clustering and parentage analyses, the studied clones had close relationship with the eight genetic groups out of the 10 used as reference. None of the local clones had genetic background of Nacional, Criollo and Curaray. A substantial number of the newly developed clones were distributed among the reference groups, particularly Marañón, Guiana, Iquitos, Trinitario, Nanay and Amelonado, suggesting their background as hybrids of these groups (Fig. 3). This observation was supported by the high proportions of admixed samples in Guiana-Marañón, Iquitos, and Amelonado-Trinitario-Nanay genetic clusters relative to Contamana-Purús genetic cluster (Fig. 1).

Figure 3. PCoA plot of 168 cacao clones in Ghana and 22 reference accessions using 45 SNP genetic data.
Parentage analysis to verify genetic purity of farmers' varieties
Parent-offspring relationships in the 752 seedlings of farmers' varieties were explored using CERVUS software. At >80% confidence level, parentage assignment identified parent-offspring trios for 48.7 to 80.3% of the seedlings sampled from six sources (Table 2). In general, 65.2% of the seedlings raised for commercial plantations had assigned parentage.
Table 2. Likelihood assignment of parentage of 752 T. cacao plants sampled from six different sources based on 58 SNP markers with LOD scores above 80% probability

Critical LOD (the natural logarithm of the likelihood) ratios for assignment of parentage are 12.45 at >95% confidence and 2.25 at >80% confidence.
Discussion
The present study analysed the population structure of recently developed cacao clones identified through rigorous testing at CRIG. In addition, the parentage of bi-clonal seedling varieties supplied to farmers for commercial plantations was verified to ascertain whether the seedlings were correctly produced. Overall, the SNP markers were informative and polymorphic, justifying their use in studying the genetic diversity, population structure and parentage of the populations under study. Further, the local clones were of diverse genetic origin, comprising eight ancestral genetic groups in contrast to four and five genetic groups associated with cacao populations in Puerto Rico (Cosme et al., Reference Cosme, Cuevas, Zhang, Oleksyk and Irish2016) and Indonesia (Dinarti et al., Reference Dinarti, Susilo, Meinhardt, Ji, Motilal, Mischke and Zhang2015), respectively. Furthermore, the study identified correct parentage for 65.2% of the bi-clonal seedlings produced in the seed gardens for commercial plantations, providing critical information for proper seed garden management. This study is among the few studies (Efombagn et al., Reference Efombagn, Sounigo, Eskes, Motamayor, Manzanares-Dauleux, Schnell and Nyassé2009; Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) that assessed the genetic purity of seedlings or clones from the cacao seed gardens.
Information content of SNP markers
The PIC, which is highly correlated with the expected heterozygosity (He) measures the informativeness of a genetic marker. In the present study, average PIC values of 0.277 and 0.256 were observed for the clones and bi-clonal seedlings, respectively. According to Botstein et al. (Reference Botstein, White, Skolnick and Davis1980), markers with PIC values < 0.25, from 0.25 to 0.5, and > 0.5 are classified as slightly informative, informative, and highly informative, respectively. However, for bi-allelic markers (such as SNPs) with maximum PIC value of 0.5, slightly informative (PIC < 0.25) and informative (0.5 > PIC > 0.25) classes of markers are applicable. Therefore, the results indicate that the SNP markers employed in this study were informative and polymorphic. The efficiency of the 45 and 58 SNPs was further revealed by the respective PID-sib values of 4.43 × 10−8 and 1.15 × 10−9, which indicate a high probability (99.999 × 10−2) of identifying an individual tree in the studied populations. The PID-sib values observed for the SNP markers are consistent with estimates reported for 44 SNPs (PID-sib = 1.0 × 10−6; Mata-Quirós et al., Reference Mata-Quirós, Arciniegas-Leal, Phillips-Mora, Meinhardt, Motilal, Mischke and Zhang2018), 53 SNPs (PID-sib = 1.0 × 10−9; Takrama et al., Reference Takrama, Ji, Meainhardt, Mischke, Opoku, Padi and Zhang2014), and 64 SNPs (PID-sib = 2.44 × 10−10; Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) used in cacao diversity studies. Furthermore, the efficiency of the two sets of SNPs used in this study corroborates the findings of Ji et al. (Reference Ji, Zhang, Motilal, Boccara, Lachenaud and Meinhardt2012) who observed that a minimum set of 26 SNPs could identify an individual cacao tree with 99.999% confidence.
Gene diversity in the cacao populations
The cacao populations had a moderate level of gene diversity (He = 0.323–0.349), which is comparable to cacao collections in Ghana (He = 0.343; Takrama et al., Reference Takrama, Ji, Meainhardt, Mischke, Opoku, Padi and Zhang2014), Honduras and Nicaragua (He = 0.367; Ji et al., Reference Ji, Zhang, Motilal, Boccara, Lachenaud and Meinhardt2012), Colombia (He = 0.314; Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017), Uganda (He = 0.332; Gopaulchan et al., Reference Gopaulchan, Motilal, Bekele, Clause, Ariko, Ejang and Umaharan2019), Dominica (He = 0.320; Gopaulchan et al., Reference Gopaulchan, Motilal, Kalloo, Mahabir, Marissa, Joseph and Umaharan2020) and north Peru (He = 0.336; Bustamante et al., Reference Bustamante, Motilal, Calderon, Mahabir and Oliva2022). The moderate genetic diversity could be explained by the large contribution of few genetic groups (limited in genetic diversity) to the cacao populations under study as well as those in the above-mentioned countries. The clones and bi-clonal seedlings were largely of three out of the 10 genetic groups identified by Motamayor et al. (Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008). Similarly, the cacao populations in Dominica, Uganda, Honduras and Nicaragua, and north Peru had significant contributions from two, three, two, and three genetic groups, respectively.
The absence of duplicates in the clonal population as revealed by pairwise multilocus matching is an indication that the ramets may have been developed from highly heterozygous bi-parental families. This observation suggests that the 168 studied clones were unique and could provide enormous genetic variation for selection of clones with improved characteristics such as bean yield, bean quality, yield efficiency, precocity, and tolerance/resistance to biotic and abiotic stresses.
Population diversity and ancestry of local clones
The delta K estimated by the Evanno's method (Evanno et al., Reference Evanno, Regnaut and Goudet2005) revealed that the cacao population used in this study could be classified into five genetic clusters. The results indicate the existence of intra-population diversity within the clonal population worthy of exploring for cacao improvement.
The present study could not identify distinct clusters for the 10 genetic groups (Motamayor et al., Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008) but classified clones of the 10 genetic groups into five clusters. This could be due to sharing of alleles by clones developed from ortets of different genetic backgrounds, resulting in joint genetic groups. Similar results were reported for cacao accessions in Colombia (Osorio-Guarín et al., Reference Osorio-Guarín, Berdugo-Cely, Coronado, Zapata, Quintero, Gallego-Sánchez and Yockteng2017) and Côte d'Ivoire (Guiraud et al., Reference Guiraud, Tahi, Fouet, Trebissou, Pokou, Rivallan, Argout, Koffi, Koné, Zoro and Lanaud2018).
Analysis of genetic structure, PCoA and parentage assignment revealed that the recently developed clones were associated with eight of the 10 genetic groups identified by Motamayor et al. (Reference Motamayor, Lachenaud, da Silva e Mota, Loor, Kuhn, Brown and Schnell2008), indicating a significant proportion of genetic variation available in the clones. However, the parentage analysis of clones further highlighted that the local clones were largely of Contamana, Marañón and Guiana genetic groups. This observation was largely influenced by the genetic composition of the newly developed clones which constituted the clonal population in this study. The background of the local clones was largely of Scavina relating to Contamana genetic group, Parinari from Marañón genetic group, and GU clones from Guiana genetic group. The absence of clones of Curaray, Nacional and Criollo genetic groups in this study agrees with classification of cacao germplasm in Ghana in previous studies (Takrama et al., Reference Takrama, Ji, Meainhardt, Mischke, Opoku, Padi and Zhang2014; Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015). The results indicate that all eight genetic groups identified in the available germplasm in Ghana (Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) have been exploited, indicating effective recurrent selection programme. The large proportion of the local clones belonging to the Contamana, Marañón and Guiana genetic groups could be explained by the strong preference for high yielding clones with large pods, large bean size, resistance to diseases and pests, precocity, establishment ease and adaptability to marginal growing conditions (Lachenaud et al., Reference Lachenaud, Oliver and Letourmy2000, Reference Lachenaud, Paulin, Ducamp and Thevenin2007; Bekele et al., Reference Bekele, Iwaro, Butler and Bidaisee2008; Paulin et al., Reference Paulin, Ducamp and Lachenaud2008; Padi et al., Reference Padi, Adu-Gyamfi, Akpertey, Arthur and Ofori2013a, Reference Padi, Ofori and Arthur2017b; Ofori et al., Reference Ofori, Padi, Assuah and Anim-Kwapong2014, Reference Ofori, Padi, Acheampong and Lowor2015, Reference Ofori, Padi and Amoako-Attah2020, Reference Ofori, Padi, Ameyaw, Dadzie, Opoku-Agyeman, Domfeh and Ansah2022). Furthermore, the Amelonado, Nanay and Iquitos background of the current farmers' varieties (Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) could explain the large selection of new clones belonging to the Contamana, Marañón and Guiana genetic groups. Further studies would be initiated to evaluate the new clones for agronomic performance to enable selection of superior clones for the seed gardens and for population improvement.
Parentage analysis to verify genetic purity of farmers' varieties
Parentage analysis was conducted to assess if bi-clonal seedlings distributed to farmers were produced correctly. Seedlings were produced through manual pollination from seed garden plots of the SPD for commercial production. The analysis revealed that 65.2% of the seedlings were assigned parent-offspring relationship, suggesting that most of the seedlings supplied to farmers for commercial plantations had their parents from the breeder's active collection or recommended seed garden clones. The 34.8% of seedlings which emerged from parents outside of the breeder's active collection could be attributed to mislabelling in seed garden clonal plots or pollen contamination from trees not belonging to the breeder's active collection. A study by Padi et al. (Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015) observed clone mislabelling within seed garden clones and among clones of the breeder's active collection in Ghana, which resulted in unexpected parentage in cacao. Besides cacao, mislabelling of parental clones and pollen contamination have been highlighted as probable causes of unexpected parentage in loblolly pine (Grattapaglia et al., Reference Grattapaglia, do Amaral Diener and dos Santos2014), Larix gmelinii (Sun et al., Reference Sun, Yu, Dong, Zhao, Wang, Zhang and Zhang2017) and coffee (Akpertey et al., Reference Akpertey, Padi, Meinhardt and Zhang2020). The mislabelling or pollen contamination, however, occurred significantly in seed garden plots meant to produce seedlings for commercial production. As erroneous labelling and pollen contamination could result in yield reduction and large variations in agronomic traits within plots of tree crop varieties (Padi et al., Reference Padi, Ofori, Takrama, Djan, Opoku, Dadzie, Bhattacharjee, Motamayor and Zhang2015; Akpertey et al., Reference Akpertey, Padi, Meinhardt and Zhang2020), it is essential to investigate clone mislabelling in all active seed gardens and rogue off-type trees in order to maintain trees with proven performance in the seed gardens to produce quality seedlings for farmers.
Conclusions
The present study assessed the genetic diversity and population structure of locally bred cacao clones to enhance effective utilization, management, and conservation of these clones. The study revealed that the local clones were distinct and had ancestral contributions from Marañón, Guiana, Contamana, Iquitos, Amelonado, Trinitario, Nanay and Purús, representing a wide range of trait diversity for cacao genetic improvement. Given that the current farmers' varieties in Ghana are composed largely of Amelonado, Nanay and Iquitos background, the new clones with different genetic backgrounds could be used to develop improved seedling varieties for farmers. However, the clones should be evaluated for their agronomic performance before being utilized in the breeding programme.
A significant proportion (34.8%) of the seedling varieties supplied to farmers for commercial plantations had incorrect parentage (possibly due to clone mislabelling and pollen contamination), which could result in underperformance and low adoption of recommended varieties. It is therefore imperative to investigate the existence of mislabelled clones or sources of pollen contamination in all active seed gardens for the appropriate measures to be applied.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1479262124000510.
Acknowledgements
The authors appreciate the support and assistance of the field and technical staff of the Plant Breeding Division of the Cocoa Research Institute of Ghana (CRIG). We are also grateful to the anonymous reviewers and editors for their valuable suggestions on the paper. This work is published with the kind permission of the Executive Director of CRIG as manuscript number CRIG/02/2024/061/002.
Authors’ contributions
Kwabena Asare Bediako: Investigation, methodology, formal analysis, writing – original draft. Francis Kwame Padi: Conceptualization, funding acquisition, project management, methodology, data curation, supervision, writing – review and editing. Ebenezer Obeng-Bio: Formal analysis, writing – review and editing. Atta Ofori: Methodology, supervision, writing – review and editing.
Funding statement
This work was supported by the Ghana Cocoa Board.
Competing interests
None.







