During the past two decades, lipid metabolism has received a great deal of attention due to the marked increase in the global prevalence of adult and childhood obesity. In this context, it is of particular interest that a range of epidemiological, clinical and animal studies have shown that maternal nutrition has a long-term influence on offspring lipid metabolism state(Reference Boney, Verma and Tucker1–Reference Sarr, Louveau and Kalbe3). It has been demonstrated that dietary protein intake during gestation and lactation is associated with offspring obesity(Reference Bol, Delattre and Reusens4–Reference Fagundes, Moura and Passos6). Although the evidence supporting a link between the fetal environment and propensity towards later lipid metabolism disorders is growing, the molecular mechanisms underlying this phenomenon are still largely unclear.
MicroRNAs (miRNA) are a class of small, endogenous, single-stranded non-coding RNA molecules that act as post-transcriptional modulators of gene expression in eukaryotes(Reference Orom, Kauppinen and Lund7). Recent studies have revealed that miRNA are crucial regulators of adipogenesis(Reference Xu, Vernooy and Guo8–Reference Mudhasani, Imbalzano and Jones10). To date, several individual miRNA have been described to regulate lipid metabolism. For example, miR-335 was reported to be closely correlated with the levels of adipocyte differentiation markers, such as PPAR-γ, adipocyte lipid-binding protein and fatty acid synthesis in 3T3-L1 adipocytes(Reference Nakanishi, Nakagawa and Tokushige11). The overexpression of miR-27 specifically inhibits adipocyte formation without affecting myogenic differentiation(Reference Lin, Gao and Alarcon12), and both miR-27a and miR-27b are negative regulators of adipogenesis(Reference Lin, Gao and Alarcon12, Reference Kim, Kim and Lee13). Knock-down of miR-378 and/or miR-378* decreases TAG accumulation(Reference Gerin, Bommer and McCoin14). Though progress has been achieved in understanding the physiological roles of miRNA in adipocytes, the roles of miRNA in mediating the effects of maternal nutrition on offspring lipid metabolism remains poorly understood. Furthermore, over 228 miRNA have been identified in pigs to date (http://www.miRbase.org), but very few of the predicted targets have been experimentally validated.
CCAAT/enhancer-binding protein-β (C/EBP-β) and PPAR-γ are well-established transcription factors involved in lipid metabolism during adipogenesis(Reference Rosen, Walkey and Puigserver15–Reference Xu, Albrecht and Viergutz18). The PPAR-γ is the master regulator of adipocyte differentiation and insulin sensitivity(Reference Yu, Wu and Cheng19). C/EBP-β is also known to directly influence adipocyte development(Reference Rosen, Hsu and Wang20). One previous study has suggested that the adipogenic differentiation of 3T3-L1 cells can be blocked via the inhibition of C/EBP-β- and PPAR-γ-dependent pathways(Reference Choi, Shin and Liu21). In the present study, we employed the Meishan sow as a model. This indigenous Chinese pig breed is famed for its high reproduction, good meat quality, resistance to roughage, high body fat deposition and slow growth. The low protein (LP) group was fed a LP diet containing 6 % protein during pregnancy followed by 7 % protein during lactation, whereas the standard protein (SP) group received 12 and 14 % protein during pregnancy and lactation, respectively. PPAR-γ and C/EBP-β were chosen as the primary genes analysed to investigate their role in regulating offspring lipid deposition. The expression levels of miRNA predicted to target these two genes were evaluated, and the functions of these candidate miRNA were verified. These results provide further evidence for a thorough understanding of the maternal effect on offspring lipid metabolism and are of clinical relevance in light of the on-going worldwide obesity epidemic.
Materials and methods
Animals and subcutaneous fat sampling
The animal experiments were carried out in the National Meishan Pig Preservation and Breeding Farm at the Jiangsu Polytechnic College of Agriculture and Forestry, Jurong, Jiangsu Province, People's Republic of China. A total of fourteen primiparous purebred Meishan gilts with an average body weight of 36·1 (sem 1·8) kg were assigned randomly into SP and LP groups. The sows in the SP group were fed diets containing 12 and 14 % crude protein, while those in the LP group were fed diets containing 6 and 7 % crude protein during gestation and lactation, respectively. The detailed nutritional compositions of these diets are shown in Table 1, and the specific feed formulations can be found in Liang et al. (Reference Liang, Zhang and Zhao22). The dietary treatments began before artificial insemination at the first observation of oestrus. Sows were fed twice daily (08.00 and 14.00 hours) and received 1·8 and 2·6 kg/d during gestation and lactation, respectively. Litter size was adjusted to seven to eight pigs per litter at 24 h post-parturition in the same group. Newborn piglets were allowed free access to their mothers and weaned at 35 d of age. One male pig per litter was killed at weaning (35 d). Subcutaneous fat tissues (with vessels and muscles removed) were obtained from the back of the neck within 20 min post-mortem, snap-frozen in liquid N2 and stored at − 80°C until further analysis.
Table 1 Nutritional components of the sow diets (percentage of original matter)

All experiments, including feeding, transport, slaughtering and sampling protocols, were undertaken following the guidelines of the Animal Ethics Committee of Nanjing Agricultural University.
RNA isolation
Total RNA was extracted from homogenised adipose tissues using the TRIzol Total RNA Kit (Invitrogen Life Technologies) and subsequently purified with the RNase-Free DNase Set (Promega) according to the manufacturer's instructions. RNA concentration was then quantified by measuring the absorbance at 260 nm in an Eppendorf BioPhotometer (Gene Company Limited). The absorption ratios (260/280 nm) of all preparations were between 1·8 and 2·0. Aliquots of 4 μg RNA were subjected to electrophoresis in a 1·4 % agarose–formaldehyde gel to verify sample integrity.
Tissue protein extraction and Western blot analysis
Total protein was extracted from 500 mg of frozen fat tissue in 1 ml lysis buffer (150 mm-NaCl, 10 mm-Tris–HCl, 5 mm-EDTA, 1 % Triton X-100 and 0·1 % SDS). The protease inhibitor cocktail (Roche Applied Science) was added according to the manufacturer's instructions. Protein concentration was measured using the Pierce BCA Protein Assay Kit (Thermo Scientific). The protein extract (40 μg) was mixed with loading buffer, denatured by boiling for 5 min and loaded on a 10 % SDS-polyacrylamide gel. After electrophoresis, proteins were transferred to nitrocellulose membranes and blocked with 3 % bovine serum albumin in 1 × Tween-Tris–buffered saline for 90 min at room temperature. After repeated washing with 1 × Tween–Tris-buffered saline, the membranes were incubated with the respective antibodies. C/EBP-β was detected using a polyclonal antibody (cs-150X, Santa Cruz Technology) at a dilution of 1:5000. C/EBP-β was detected at approximately 38 kDa. The PPAR-γ antibody (Bioworld Technology) was used at a dilution of 1:500. A protein band at approximately 57 kDa was observed. An antibody against β-actin (Kangcheng, diluted 1:500) was used as internal standard. Goat anti-rabbit IgG peroxidase-conjugated secondary antibodies (Bioworld Technology) were used at a dilution of 1:5000. Finally, the membrane was washed and the specific signals were detected by chemiluminescence using the LumiGlo substrate (SuperSignal West-Pico Trial Kit, Pierce). The C/EBP-β and PPAR-γ contents of the samples are given as the band density values of C/EBP-β and PPAR-γ relative to β-actin. Band densities were analysed with Versa Doc™ 4000 MP (Bio-Rad).
Bioinformatics methods
To assay the deregulation of the miRNA targets of differentially expressed target genes, we used four of the leading miRNA target prediction algorithms: miRanda (http://microrna.sanger.ac.uk/sequences/), PicTar (http://pictar.mdc-berlin.de/)(Reference Lewis, Burge and Bartel23), TargetScan (release 5.1, http://www.targetscan.org/)(Reference John, Enright and Aravin24) and miRGen (http://www.diana.pcbi.upenn.edu/miRGen.html)(Reference Krek, Grun and Poy25).
MicroRNA RT-PCR quantification
The RT-PCR analysis of miRNA expression was performed in an Mx3000P system (Stratagene) with specific primers (Table 2). Briefly, total RNA was extracted from adipocytes using the TRIzol reagent (Invitrogen) and subsequently purified with the RNase-Free DNase Kit (Promega) according to the manufacturer's instructions. Then, miRNA was treated with the Poly(A) Tailing Kit (Ambion, AM1350) to add a poly-A tail to the 3′ end of each RNA transcript. The tailing reactions contained 4 μg RNA samples (1 μg/μl), 4 μl of 5 × Escherichia coli poly (A) polymerase (E-PAP) buffer, 2 μl of 25 mm-MgCl2, 2 μl of 10 mm-ATP, 0·8 μl E-PAP (2 U/μl) and 2 pmol exogenous control 5 (E5), adjusted to 20 μl with nuclease-free water. The 20 μl reactions were incubated for 1 h at 37°C and held at 4°C. Then, the sample was purified to remove any residual tailing reagents. Complementary DNA were synthesised from the tailed RNA using gene-specific primers with oligo-dT (a short sequence of deoxy-thymine nucleotides) adapters. RT reactions contained 2 μg poly-A-tailed miRNA, 1 μl oligo-dT adapter (1 μg/μl) and nuclease-free water. The 10 μl reactions were incubated for 5 min at 70°C (RT1). The RT2 reactions consisted of the entire RT1 reactions, mixed with 5 μl Moloney murine leukaemia virus reverse transcriptase (M-MLV) 5 × buffer (containing 250 mm (pH 8·3) Tris–HCl, 15 mm-MgCl2, 375 mm-KCl and 50 mm-dithiothreitol), 1·25 μl of 10 mm-deoxyribonucleotide triphosphate, 1 μl M-MLV RNase (200 U/μl) and 0·5 μl RNase inhibitor (40 U/μl). The 25 μl reactions were incubated at 42°C for 1 h and then at 95°C for 5 min. The 25 μl PCR mixture included 2 μl RT product, 2 μl primers, 8·5 μl sterile 3 d H2O and 12·5 μl SYBR Premix Ex Taq TM (TaKaRa). PCR were run on an Mx3000P instrument (Agilent Technologies) and analysed using Mx3000P System SDS software (Stratagene).
Table 2 The primer sequences of the putative microRNA (miRNA) for RT-PCR

To evaluate miRNA expression, the E5 small nuclear RNA was used as an exogenous control to normalise the RNA input. E5 is designed as a random sequence and its length is equal to that of the target miRNA (approximately 22 nt); its sequence after synthesis does not yield any matching sequences when blasted to genomic DNA. Furthermore, when the synthesised sequence was used as upstream primer and universal downstream primer to PCR, no Gershgorim band was amplified. The C t value is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. The fold change was calculated using the 2−ΔΔC t method. All experiments were carried out in triplicate.
Plasmid construction
Pig genomic fragments of miR-130b, miR-374b and precursors of approximately 89 bp (Table 3) were synthesised by Invitrogen. There is only one predicted conserved target site for miR-130b in the entire 3′-untranslated region (UTR) of PPAR-γ (http://www.targetscan.org) and one predicted conserved target site for miR-374b in the entire 3′-UTR of C/EBP-β (http://www.targetscan.org). A 386 bp fragment of the PPAR-γ 3′-UTR was amplified by PCR using the primers 5′-GCTGCTGCAAGTAATAAAG-3′ and 5′-TAAAGGAAGGAAGAGGGAG-3′, and a 389 bp fragment of C/EBP-β 3′-UTR was amplified by PCR using the primers 5′-CCACAGTGACTCCGGGAAG-3′ and 5′-CGTAGGAACATCTTTAAGCGA-3′. The 386 bp fragment, which contains a broadly conserved motif in the vertebrates for miR-130b (http://www.targetscan.org), and 389 bp fragment, which contains a broadly conserved motif in the vertebrates for miR-374b (http://www.targetscan.org), were cloned downstream of the luciferase gene in the pGL3-Control report luciferase vector (Promega). These constructs, named pGL3-Control/PPAR-γ and pGL3-Control/C/EBP-β, were transfected into HeLa cells. The PCR products were subcloned into the luciferase reporter pGL3-Control using XbaI (Invitrogen). Precursor miR-130b and miR-374b were annealed using annealing buffer (5 × ), miRNA precursor upstream sequence (50 μm) and downstream sequence (50 μm). The 50 μl reaction mixtures were incubated in a ninety-six-well optical plate at 95°C for 2 min, then subjected to touchdown PCR (decrease 0·1°C/8 s till 25°C); the PCR products were subcloned into the pSilencer 3.0-H1 small interfering RNA expression vector using BamHI and Hind III (Invitrogen).
Table 3 Primer sequences of precursor microRNA

ssc-mir-130b, Sus scrofa mir-130b; ssc-mir-374b, Sus scrofa mir-374b; ssc-miR-SC, Sus scrofa miRNA scrambled control; F, forward; R, reverse.
DNA transfection
Approximately 3 × 104/cm3 HeLa cells were seeded and cultured in 25 cm2 cell culture bottles. When the cells reached 90–95 % confluence, they were digested using 0·25 % trypsin and cotransfected with 100 ng pGL3-Control/PPAR-γ 3′-UTR fluorescent luciferase reporter plasmid, 10 ng HSV-thymidine kinase promotor (pRL-TK) plasmid (used to normalise for transformation efficiency), 100 ng pSilence 3.1-H1 neo miR-130b and 100 ng pGL3-Control/C/EBP-β 3′-UTR fluorescent luciferase reporter plasmid, 10 ng pRL-TK plasmid or 100 ng pSilence 3.1 H1-neo miR-374b, with an electroporation device. Negative controls were cotransfected with 100 ng miR-SC, 100 ng target genes 3′-UTR fluorescent luciferase reporter plasmid and 10 ng pRL-TK plasmid. After transfection, the cells were counted. The cell density was approximately 2 × 104 cells/cm3. The transfected HeLa cells were incubated at 5 % CO2 and 37°C for 24 and 48 h, respectively.
Dual luciferase activity assay
At 24 and 48 h after transfection, firefly and renilla luciferase activity were measured using a Dual-Luciferase Assay Kit (Promega) with a plate reader (Perkin Elmer). The renilla and firefly luciferase signals were detected using the Veritas Microplate Luminometer (Turner Biosystems). The firefly luciferase signal was normalised to the renilla luciferase signal. The normalised firefly luciferase activity was compared between miR-130b and the miR-SC cells using one-way ANOVA. The results were expressed as relative activity. Each target construct was tested in triplicate, and the assay was repeated to confirm the results.
Statistical analysis
All data are presented as means with their standard errors. Statistical analyses were carried out with Statistical Program for Social Sciences software 17.0 for Windows (SPSS, Inc.). Differences were tested with ANOVA using a t test for independent samples. P values of less than 0·05 were considered significant.
Results
Offspring body weight and serum NEFA concentration
As shown in Table 2, body weight was significantly lower in the LP groups than the SP groups (P <0·05) at weaning. Moreover, the greatest difference in the back fat thickness (P< 0·05) of offspring between the LP and SP groups was detected at the weaning stage (Table 4).
Table 4 Body weight, back fat thickness, serum TAG and NEFA concentration in offspring weaned piglets (Mean values with their standard errors, n 7)

SP, standard protein; LP, low protein.
* Mean value was significantly different from that of the SP group (P <0·05).
Maternal dietary protein restriction had no significant effect on serum TAG concentrations. Serum NEFA concentration in the LP group was reduced, albeit not significantly (P= 0·066), compared with the SP group (Table 4).
PPAR-γ and CCAAT/enhancer-binding protein-β protein expression in subcutaneous fat
The protein expression of PPAR-γ (P< 0·05; Fig. 1(a)) was significantly decreased in the subcutaneous fat of offspring pigs at weaning in the LP group compared to the SP group; meanwhile, C/EBP-β protein levels tended to be decreased (P= 0·056) in the subcutaneous fat of offspring pigs in the LP group (Fig. 1(b)).
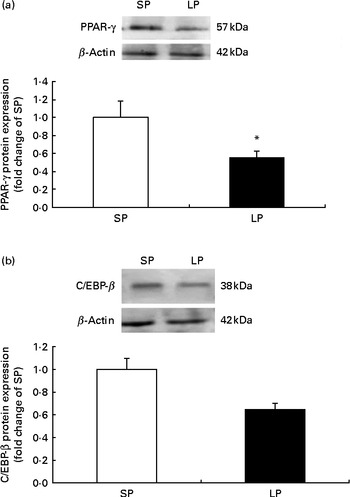
Fig. 1 Effect of maternal dietary protein on (a) PPAR-γ and (b) CCAAT/enhancer-binding protein-β (C/EBP-β) protein expression in the subcutaneous fat of piglets at weaning age. SP, maternal standard protein diet. LP, maternal low-protein diet. Values are means, with their standard errors represented by vertical bars (n 6). * Mean value was significantly different from that of the SP group (P< 0·05).
MicroRNA expression level targeting the PPAR-γ and CCAAT/enhancer-binding protein-β
As shown in Fig. 2, among the miRNA predicted to target the PPAR-γ gene, maternal low dietary protein significantly induced the expression of miR-130b (P< 0·05; Fig. 2(a)), but miR-128, 130a, 27a/b and 301 were expressed at similar levels in both groups. Among the miRNA predicted to target the C/EBP-β gene, maternal low dietary protein significantly induced the expression of miR-374b, and miR-155, 374a, 362, 191, 455 and 423 did not vary significantly (P>0·05) between the LP and SP groups (Fig. 2(b)).

Fig. 2 Effect of maternal dietary protein on microRNA (miRNA) expression in the subcutaneous fat of piglets at weaning age. (a) miRNA targeting PPAR-γ. (b) miRNA targeting CCAAT/enhancer-binding protein-β. SP (□), Maternal standard protein diet; LP (■), maternal low protein diet. Values are means, with their standard errors represented by vertical bars (n 6). * Mean value was significantly different from that of the SP group (P< 0·05).
Validation of ssc-miR-130b (Sus scrofa miR-130b) targeting PPAR-γ 3′-untranslated region and ssc-miR-374b (Sus scrofa miR-374b) targeting CCAAT/enhancer-binding protein-β 3′-untranslated region
The miR-130b had highly conserved sites for binding to the 3′-UTR of PPAR-γ, and miRNA-374b had highly conserved sites and poorly conserved sites for binding to the 3′-UTR of the 3′-UTR of C/EBP-β (Fig. 3). To ascertain whether miR-130b is able to recognise the PPAR-γ 3′-UTR or miRNA-374b is able to recognise the C/EBP-β 3′-UTR, we generated a luciferase reporter DNA construct containing the 387 bp pig PPAR-γ 3′-UTR with a putative miR-130b binding site and an ssc-miR-130b overexpression plasmid. The C/EBP-β 3′-UTR and ssc-miR-374b overexpression plasmids were constructed and analysed in the same way. When the pGL3-Control/PPAR-γ 3′-UTR fluorescent luciferase reporter plasmid and miR-130b overexpression vector were cotransfected into HeLa cells, luciferase activity was significantly suppressed by the ectopic expression of miR-130b after cotransfection for 24 or 48 h. Similar results were found for the cotransfection of the miR-374b overexpression vector and the luciferase reporter containing the C/EBP-β 3′-UTR (Fig. 4).

Fig. 3 The miR-130b target site in the 3′-untranslated region (UTR) of PPAR-γ and the miR-374b target site in the 3′-UTR of CCAAT/enhancer-binding protein-β (C/EBP-β). (a) The single predicted binding site of miR-130b in the 3′-UTR of human PPAR-γ. (b) The predicted conserved binding site of miR-374b in the 3′-UTR of pig C/EBP-β. (c) The predicted poorly conserved binding site of miR-374b in the 3′-UTR of pig C/EBP-β.

Fig. 4 Validation of (a) ssc-miR-130b targeting of the PPAR-γ 3′-untranslated region (UTR) and (b) ssc-miR-374b targeting of the CCAAT/enhancer-binding protein-β 3′-UTR at 24 and 48 h of transfection. Values are means, with their standard errors represented by vertical bars (n 3). * Mean value was significantly different from that of miRNA scrambled control (P< 0·05). (a) □, miRNA scrambled control; ■, miRNA-130b. (b) □, miR scrambled control; ■, miR-374b.
Discussion
Many previous studies have demonstrated that maternal protein level during pregnancy and/or lactation can affect the body composition of her offspring later in life(Reference Bol, Delattre and Reusens4–Reference Fagundes, Moura and Passos6). Consistent with the results of previous studies(Reference Plagemann, Waas and Harder26–Reference Woods, Ingelfinger and Nyengaard29), the present study demonstrated a decrease in body weight of offspring subjected to maternal protein restriction during pregnancy and lactation at weaning age. Early life is a critical period for body lipid deposition(Reference Wells30). Previous reports on the effects of maternal protein restriction on plasma parameters are inconsistent. Lucas et al. (Reference Lucas, Baker and Desai31) showed that the offspring of protein-restricted mothers exhibited a long-term reduction in plasma cholesterol, HDL-cholesterol and TAG concentrations compared with controls, whereas Desai et al.(Reference Desai, Byrne and Meeran32), Zambrano et al.(Reference Zambrano, Bautista and Deas28) and Qasem et al.(Reference Qasem, Cherala and D'Mello33) reported that low maternal protein consumption has no significant influence on offspring plasma lipid metabolism indices. In the present study, though the serum NEFA concentration in LP group appeared to be reduced slightly (P= 0·066) and the serum TAG levels did not differ significantly between the two groups. Nevertheless, the piglets in the maternal LP group demonstrated significantly decreased subcutaneous fat at the end of the suckling period.
Many transcriptional factors are involved in adipocyte differentiation and lipid metabolism. Previous studies have shown that the overexpression of PPAR-γ can promote fat deposition(Reference Rosen, Sarraf and Troy34, Reference Barak, Nelson and Ong35). Compared with wild-type mice, PPAR-γ-deficient mice exhibited smaller adipocytes and decreased fat mass(Reference Kubota, Terauchi and Miki36). Members of the C/EBP family are also important for adipogenesis, and C/EBP-β contributes to the transcriptional activation of PPAR-γ in early adipogenesis(Reference Lefterova, Zhang and Steger37). In the present study, we found that the mRNA expression of the key transcription factors PPAR-γ and C/EBP-β was not significantly different between the two groups(Reference Liang, Zhang and Zhao22). However, when the protein levels of these two genes were measured, the protein level of PPAR-γ was significantly decreased and C/EBP-β was not significantly decreased (P= 0·056) in the subcutaneous fat of offspring pigs in the LP group. The incongruity between the mRNA and protein levels hinted that post-transcriptional mechanisms might play a role in regulating these two key lipogenesis factors.
The miRNA are strong post-transcriptional regulators of mammalian differentiation. Recent bioinformatics predictions of miRNA targets in vertebrates demonstrated that hundreds of miRNA are involved in regulating up to 30 % of human protein-coding genes, though miRNA comprise less than 1 % of all predicted genes in human subjects(Reference Lewis, Burge and Bartel23, Reference Krek, Grun and Poy25). Further studies have indicated that miRNA are involved in the regulation of many biological processes, including fat metabolism(Reference Xu, Vernooy and Guo8). Although several miRNA have been shown to affect the regulation of adipocyte development, these results have mainly been acquired from cell culture models in vitro (Reference Romao, Jin and Dodson17). The roles of miRNA in adipose tissue, and their maternal effects in particular, are largely unknown. To our knowledge, the present study is the first to report a difference in the expression of miRNA targeting the PPAR-γ and C/EBP-β in the adipose tissue of offspring of mothers fed a LP diet during gestation and lactation.
We identified six candidate PPAR-γ-targeting miRNA by bioinformatics analyses. However, only miRNA-130b expression was significantly different between the LP and SP groups. The miR-130 has been shown to impair adipogenesis upon overexpression by targeting both the coding region and 3′-UTR of the PPAR-γ mRNA in the 3T3-L1 cell line(Reference Lee, Lee and Abdelmohsen38). The miR-130 can be expressed in two forms, miR-130a and miR-130b; however, in the present study, only miR-130b expression was significantly up-regulated. Target prediction algorithms identify a single miR-130b binding site in the 3′UTR of PPAR-γ, and the PPAR-γ 3′UTR is highly conserved among mammals. Luciferase reporter assays were performed to fully validate the predicted miRNA–mRNA interactions. When a miR-130b overexpression vector was cotransfected into HeLa cells with a luciferase reporter vector containing PPAR-γ 3′-UTR, luciferase activity was significantly suppressed. These results confirmed that miR-130b directly recognises and binds to the 3′-UTR of PPAR-γ, thereby suppressing PPAR-γ gene expression. In a previous study, miR-27 was shown to repress PPAR-γ in human multi-potent adipose-derived stem cells(Reference Karbiener, Fischer and Nowitsch39). However, in the present study, neither miR-27a nor miR-27b demonstrated a significant difference between the two groups.
Among the miRNA related to C/EBP-β, miR-378/378*(Reference Gerin, Bommer and McCoin14) and miR-143(Reference Esau, Kang and Peralta40, Reference Xie, Lim and Lodish41) have been reported to influence C/EBP-β transcriptional activity, but there is no direct evidence to confirm that these miRNA target C/EBP-β in the context of lipid metabolism. In macrophage and B-cell studies, miR-155 was shown to directly target C/EBP-β(Reference Worm, Stenvang and Petri42, Reference Costinean, Sandhu and Pedersen43). The present study demonstrates for the first time that miR-374b expression is significantly different between the SP and LP groups. The miRNA-374 expression has been reported to vary in association with lung cancer(Reference Miko, Czimmerer and Csanky44). Furthermore, in the present study, miR-374b overexpression was able to reduce the activity of a luciferase reporter containing the C/EBP-β 3′-UTR after cotransfection for 24 or 48 h. These results indicate that miR-374b can directly recognise and bind to the 3′-UTR of C/EBP-β and suppress C/EBP-β expression.
In contrast, miR-130b did not alter the activity of a luciferase reporter that has no PPAR-γ 3′-UTR, and miR-374b did not alter the activity of a luciferase reporter that has no C/EBP-β 3′-UTR (data not shown). These results imply that inhibitory effects of miR-130b on PPAR-γ expression and miR-374b on C/EBP-β expression are quite selective, as suggested by bioinformatics analyses. The present results indicate that miRNA-130b and miR-374b are likely to interact with the 3′-UTR of PPAR-γ and C/EBP-β, respectively, and consequently down-regulate their expression at the post-transcriptional level. In addition, there are two predicted miR-374b binding sites in the C/EBP-β 3′-UTR. One is a conserved binding site between positions 484 and 490, and the other site is a poorly conserved motif between positions 154 and 160. In the present study, we focused on the highly conserved binding site for the validation of miR-374b.
In conclusion, the present data represent the first description of the post-transcriptional regulation of lipid metabolism in the weaning-age offspring of female pigs fed a LP diet. Among several predicted miRNA tested, we provide evidence that miR-130b and miR-374b may play a role in lipid metabolism regulation. Anyway, every miRNA is thought to regulate an average of approximately 200 target genes and have widespread effects(Reference Krek, Grun and Poy25). A complete understanding of the biological functions of miR-130b and miR-374b in lipid metabolism requires further studies.
Acknowledgements
The present study was supported by the National Basic Research Program of China (2012CB124703), the Special Fund for Agro-scientific Research in the Public Interest (201003011), the Major National Science and Technology Projects of China (2009ZX08009-138B) and the Priority Academic Program Development of Jiangsu Higher Education Institutions. X. Y. and R. Z. designed the study; S. P. and Y. Z. performed the experiments; S. P. and X. Y. analysed the data and wrote the paper. The authors declare that there is no conflict of interest.










