Acetolactate synthase (ALS) catalyzes one of the vital steps in the biosynthesis of the branched-chain amino acids valine, leucine, and isoleucine in plants and microorganisms (Dailey and Cronan Reference Dailey and Cronan1986; Shaner Reference Shaner1991). ALS is the target site of several herbicide classes, such as sulfonylureas (SUs), imidazolinones (IMIs), triazolopyrimidines (TPs), pyrimidinylthiobenzoates (PTBs), and sulfonylaminocarbonyltriazolinones (SCTs). Differential selectivity of ALS inhibitors enabled their widespread use in many crops, including wheat (Triticum aestivum L.), cotton (Gossypium hirsutum L.), soybean [Glycine max (L.) Merr.], and corn (Zea mays L.) to control a broad spectrum of grass and broadleaf weeds (Beyer et al. Reference Beyer, Duffy, Hay and Schlueter1988). ALS inhibitors are also popular because of their low use rates, high efficacy, low mammalian toxicity, and cost-effectiveness. Extensive and widespread use of these herbicides created a strong selection pressure, resulting in the evolution of herbicide resistance in a number of weed species. SU herbicide resistance was reported as early as 1987 in prickly lettuce (Lactuca serriola L.) in no-till winter wheat in Idaho (Mallory-Smith et al. Reference Mallory-Smith, Thill and Dial1990), and since then 159 weed species have evolved resistance to ALS inhibitors, including many Amaranthus species (Heap Reference Heap2017).
Palmer amaranth has evolved resistance to a number of widely used herbicides with different herbicide sites of action, including ALS, microtubule, photosystem II, 5-enolpyruvylshikimate-3-phosphate synthase, hydroxyphenylpyruvate dioxygenase, and protoporphyyrinogen inhibitors across United States (Heap Reference Heap2017). The most commonly reported mechanism conferring resistance to ALS inhibitors in weed species is target site–based, due to point mutations resulting in single amino acid substitutions in the ALS gene (Shaner Reference Shaner1991). Predominantly, a high level of resistance is bestowed by these alterations, because of reduced sensitivity of the target enzyme to the herbicides. The number of mutations detected that lead to target-site alterations has increased dramatically in last decade, and a total of 26 amino acid substitutions were identified across 8 amino acid positions (Tranel et al. Reference Tranel, Wright and Heap2016). The Pro-197 amino acid position on the ALS gene is the most commonly reported point mutation in resistant populations. So far, 11 substitutions at this position have been reported in ALS inhibitor–resistant weeds such as giant ragweed (Ambrosia trifida L.), kochia [Kochia scoparia (L.) Schrad.], and many Amaranthus populations, e.g., tall waterhemp [Amaranthus tuberculatus (Moq.) Sauer], redroot piweed (Amaranthus retroflexus L.), and smooth pigweed (Amaranthus hybridus L.) (Burgos et al. 2001; Foes et al. Reference Foes, Liu, Vigue, Stoller, Wax and Tranel1999; Guttieri et al. Reference Guttieri, Eberlein and Thill1995; Patzoldt and Tranel Reference Patzoldt and Tranel2002; Varanasi et al. Reference Varanasi, Godar, Currie, Dille, Thompson, Stahlman and Jugulam2015).
Non–target site resistance, endowed by metabolism of ALS inhibitors, was also identified in many ALS inhibitor–resistant grasses such as rigid ryegrass (Lolium rigidum Gaudin) and broadleaf weeds like wild mustard (Sinapsis arvensis L.) and tall waterhemp (Christopher et al. Reference Christopher, Powles, Liljegren and Holtum1991; Cotterman and Saari Reference Cotterman and Saari1992; Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015; Veldhuis et al. Reference Veldhuis, Hall, O’Donovan, Dyer and Hall2000). Ryegrass populations (SLR31 and VLR69) from Australia that are resistant to multiple herbicides showed metabolism-based cross-resistance to SU (e.g., chlorsulfuron) and IMI (e.g., imazamethabenz) herbicides (Christopher et al. Reference Christopher, Powles, Liljegren and Holtum1991; Heap and Knight Reference Heap and Knight1986). The non–target site based mechanism of resistance to chlorsulfuron has been reported to be due to cytochrome P450 monooxygenase activity (Christopher et al. Reference Christopher, Preston and Powles1994). Natural tolerance and selectivity to ALS inhibitors in crops such as wheat, barley (Hordeum vulgare L.), rice (Oryza sativa L.), corn, and soybeans is also bestowed because of rapid detoxification of the ALS inhibitors via a wide array of cytochrome P450 enzymes (Brown Reference Brown1990).
ALS inhibitor–resistant Palmer amaranth was first documented in Kansas in 1993 (Horak and Peterson Reference Horak and Peterson1995) and later reported in many states across the United States. However, the precise mechanism of resistance is not known. This research was based on the hypothesis that target site–based or non–target site based mechanisms may contribute to resistance to ALS inhibitors in Palmer amaranth. The objective of this study was to determine the molecular basis of chlorsulfuron resistance in Palmer amaranth from Kansas and investigate this population’s cross-resistance to other chemical classes of ALS inhibitors.
Materials and Methods
Plant Material and Growth Conditions
A chlorsulfuron-resistant Palmer amaranth population collected from a field in Stafford County, KS (Thompson et al. Reference Thompson, Peterson and Lally2012) was designated as KSR, and two susceptible populations from KS (KSS) and Mississippi (MSS) were used for comparison. Seeds of KSR, KSS, and MSS Palmer amaranth were germinated in small trays (25 by 15 by 2.5 cm) with commercial potting mixture (Miracle-Gro®, Marysville, OH) and transplanted into small pots (6 by 6 by 6.5 cm) when seedlings were 2- to 3-cm tall under greenhouse conditions (25/20 C day/night temperature and 15/9-h photoperiod, supplemented with additional 250 µmol m−2 s−1 illumination provided with sodium-vapor lamps). Palmer amaranth plants (5- to 6-cm tall) were transferred to a growth chamber maintained at 32.5/22.5 C, 15/9-h photoperiod, 60% to 70% relative humidity. Light in the growth chamber was provided by fluorescent bulbs delivering 550 µmol m−2 s−1 photon flux at plant canopy level. Plants were regularly watered as needed under both greenhouse and growth chamber conditions.
Dose–Response Assay
The populations of KSS, MSS, and KSR Palmer amaranth were grown as described, and 10- to 12-cm-tall plants were treated with the field recommended rate of chlorsulfuron (Glean® XP, DuPont Crop Protection, 18 g ai ha−1; Table 1) to determine the frequency of resistant individuals in the KSR population before conducting the dose–response assay. It was found that approximately 70% to 75% of plants survived the treatment. This suggests that the KSR population was segregating for chlorsulfuron resistance or susceptibility (unpublished data). For the dose–response analysis, 10- to 12-cm-tall Palmer amaranth plants at the 8- to 10-leaf stage were treated separately with the following doses: 0, 0.56, 1.12, 2.25, 4.5, 9, 18 (1X), 36, 72, 108, and 144 g ha−1, where 1X represents the field rate of chlorsulfuron. A nonionic surfactant (NIS) at 0.25% v/v was used as an adjuvant in all the treatments. Herbicide treatments were applied with a bench-type sprayer (Research Track Sprayer, Generation III, De Vries Manufacturing, RR 1 Box 184, Hollandale, MN) equipped with a flat-fan nozzle tip (80015LP TeeJet® tip, TeeJet Spraying Systems, P.O. Box 7900, Wheaton, IL) delivering 168Lha−1 at 222 kPa in a single pass at 4.8 km h−1. After treatment, plants were returned to the growth chamber and maintained under the conditions described earlier. Treatments were arranged in a completely randomized design with six replications, and the experiment was repeated three times. Aboveground biomass was harvested 3 weeks after treatment (WAT) and plants packed in paper bags were oven-dried at 60 C for a week before dry biomass was measured.
Table 1 List of herbicides used in the study

DNA Isolation and ALS Gene Sequencing
Fresh leaf tissue was collected from KSR plants that survived different doses of chlorsulfuron in whole-plant dose–response experiments and also untreated susceptible (KSS and MSS) Palmer amaranth. The collected tissue (100 mg) was frozen in liquid nitrogen, and genomic DNA (gDNA) was extracted using the DNeasy Plant Mini Kit (Qiagen) according to the manufacturer’s instructions. DNA was quantified using a Nanodrop® 1000 (Thermo Scientific), and quality was analyzed using 0.8% agraose gel electrophoresis. Forward (ALSF: 5′-TCCTCGCCGCCCTCTTCAAATC-3′) and reverse (ALSR: 5′-CAGCTAAACGAGAGAACGGCCAG-3′) primers were used to amplify the ALS gene of ~2,000 bp in length (GenBank Accession U55852; Whaley et al. Reference Whaley, Wilson and Westwood2007). PCR reactions were performed using T100TM Thermal Cycler (Bio-Rad, Hercules, CA). Each PCR reaction contained 80 to 100 ηg of gDNA, 0.5 µM of forward and reverse primers each, and PCR Master Mix (Promega). The PCR product was purified using the GENEjet PCR purification kit (Thermo Fisher Scientific) according to the manufacturer’s instructions and was sequenced using the ALSF and ALSR primers at the GENEWIZ sequencing facility (South Plainfield, NJ). Alignment of DNA sequences from KSR, MSS, and KSS was performed using MultAlin software (Corpet Reference Corpet1988).
Effect of Malathion Pretreatment on Chlorsulfuron Resistance
Malathion, a cytochrome P450 inhibitor pretreatment, was used to investigate its ability to reverse the chlorsulfuron resistance. KSR Palmer amaranth plants 10- to 12-cm tall were treated with malathion as described in Ma et al. Reference Ma, Kaundun, Tranel, Riggins, McGinness, Hager, Hawkes, McIndoe and Riechers2013. Malathion at a rate of 2000 g ai ha−1 with 0.25% NIS as an adjuvant was applied 1 h before chlorsulfuron application. Two days after herbicide treatment, 50 ml pot−1 of 5 mM malathion solution was used to drench the soil. Untreated plants and plants treated only with malathion were used as controls to compare the efficacy of malathion in increasing the sensitivity of the KSR plants to the herbicide.
Screening for Cross-resistance
The KSR and MSS Palmer amaranth seedlings were grown under the greenhouse conditions described earlier. To determine cross-resistance of KSR to other classes of ALS inhibitors, the plants were treated separately with field recommended rates of thifensulfuron, 36 g ai ha−1 (Harmony®, DuPont Crop Protection); imazamox, 35 g ai ha−1 (Beyond®, BASF Ag Products); propoxycarbazone, 44 g ai ha−1 (Olympus®, Bayer CropScience) and pyrithiobac, 73 g ai ha−1 (Staple®, DuPont Crop Protection), which belong to the SU, IMI, SCT, and PTB classes, respectively (Table 1). All treatments included NIS at 0.25% v/v as an adjuvant and sprayed using the bench-type sprayer described earlier. Plant survival was assessed 3 to 4 WAT to determine the resistance of KSR Palmer amaranth to each herbicide.
Statistical Analysis
Biomass data were subjected to ANOVA in R v. 3.1.2 to test for treatment by experiment interaction, and the means were compared using Tukey’s honest significant difference test. The data from repeated experiments were pooled, because the interaction of herbicide treatment and experiment was not significant (P>0.05).
Dose–response data (expressed as a percentage of the untreated control) were analyzed using the ‘drc’ package in R v. 3.1.2 (Ritz and Streibig 2014; Seefeldt et al. Reference Seefeldt, Jensen and Fuerst1995). The three-parameter log-logistic model in Equation 1 was used to show the relationship between herbicide rate and biomass:
where Y is the response (dry biomass or plant health), expressed as percentage of the untreated control; d is asymptotic value of Y at the upper limit; b is the slope of the curve around GR50 (the herbicide rate giving response halfway between d and the lower asymptotic limit, which was set to 0); and x is the herbicide rate. Resistance index (R/S) was calculated as the ratio of the GR50 of KSS and KSR populations.
Malathion data of KSR were analyzed using ANOVA, and the means were compared using Tukey-Kramer’s pairwise comparison test at P=0.05. Palmer amaranth biomass data were converted into percent biomass reduction compared with the untreated control using the following formula (Chahal et al. Reference Chahal, Aulakh, Rosenbaum and Jhala2015): Biomass reduction (%)=[(C−B)/C]×100, where C is the mean biomass of the untreated control and B is the mean biomass collected from treatments. Expected value for herbicide interactions was calculated with the use of the Colby equation (Colby Reference Colby1967): E=(X+Y)−(XY/100), where E is expected biomass reduction (%) with malathion+chlorsulfuron and X and Y are observed biomass reduction (%) with malathion and chlorsulfuron, respectively. The expected and observed control values for herbicide combination were subjected to a t test to determine whether means were different. The herbicide combination was considered antagonistic if the expected mean was significantly greater than the observed mean. If the expected mean was significantly lower than the observed mean, the herbicide combination was considered synergistic.
Results and Discussion
Chlorsulfuron Dose–Response Assay
The result of the dose–response assay confirmed a very high level of resistance to chlorsulfuron in the KSR population when compared with MSS or KSS Palmer amaranth (Figure 1). The shoot dry weight relative to the untreated control measured at 3 WAT gradually decreased with increasing rate of chlorsulfuron. However, the KSR plants survived 8X the field rate of chlorsulfuron (Figure 1C). The MSS or KSS Palmer amaranth responded differently to chlorsulfuron application. The MSS population showed a high level of sensitivity and was not able to survive even the lowest dose used (0.56 g ai ha−1), which is 32X less than the field rate (Figure 1A). On the other hand, the KSS population survived one-fourth of the field recommended dose, but was completely killed at half of the field rate (Figure 1B). The ALS inhibitors are widely known for their efficacy at very low doses with high crop selectivity in controlling many broadleaf and grass weeds (Mazur and Falco Reference Mazur and Falco1989; Ray Reference Ray1984). The differential sensitivity in the two susceptible populations might be due to inherent genetic variation and also might be due to the environmental adaptations. Because the MSS population was completely killed at the lowest dose (0.56 g ha−1) of chlorsulfuron used, the dose–response curve could not be fit to analyze the GR50 value with KSS and KSR (Figures 1A and 2). The amount of chlorsulfuron required to reduce Palmer amaranth growth to 50% (GR50) at 3 WAT was 1.08 g ha−1 for KSS, while the GR50 of KSR was 303 g ha−1 (Figure 2; Table 2). Overall, the KSR population was more than 275 times more resistant to chlorsulfuron relative to the KSS Palmer amaranth (Figure 2). In comparison, another Amaranthus species, redroot pigweed, was more than 429 and 34 times more resistant to thifensulfuron and nicosulfuron, other commonly used SU herbicides (Scarabel et al. 2007). In another whole-plant dose–response study, Powell amaranth (Amaranthus powellii S. Wats.) and redroot pigweed were 1,257 and 270 times more resistant to thifensulfuron when compared with their susceptible controls (Ferguson et al. 2001).
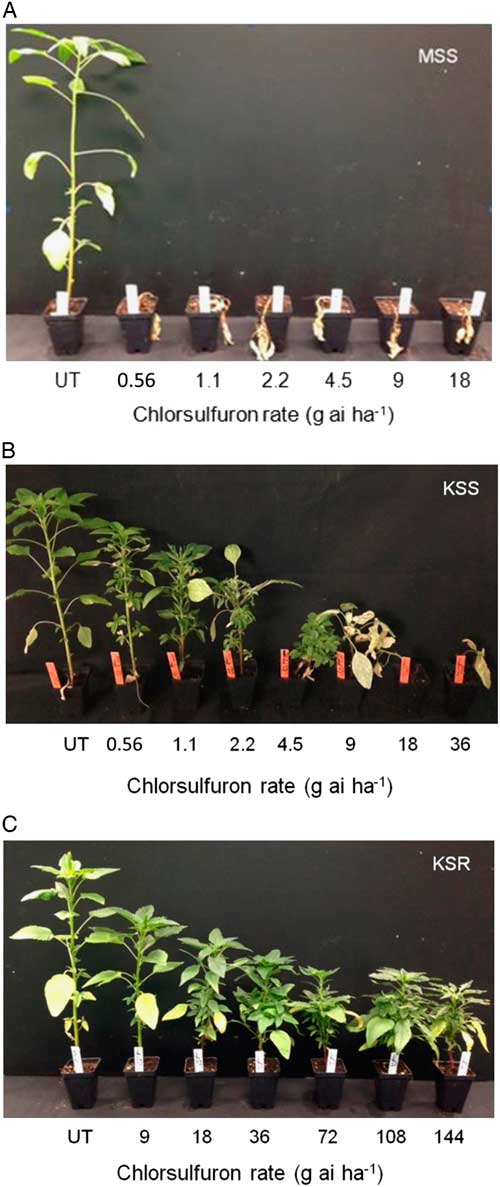
Figure 1 Whole-plant response of susceptible and resistant Palmer amaranth populations to different doses of chlorsulfuron at 3 WAT: (A) MSS, (B) KSS, and (C) KSR. UT, untreated.

Figure 2 Nonlinear regression analysis of aboveground dry biomass of KSS and KSR Palmer amaranth to different doses of chlorsulfuron at 3 WAT. Symbols represent averages of 12 to 13 replicates fit with a three-parameter log-logistic model; model parameters are shown in Table 2.
Table 2 Summary parameters describing the response of KSS (susceptible) and KSR (resistant) Palmer amaranth aboveground dry biomass to rates of chlorsulfuron at 3 WAT.Footnote a, Footnote b

a Abbreviations: WAT, weeks after treatment; b, relative slope around GR50; d, upper limit of the response; GR50, chlorsulfuron rate causing 50% reduction in aboveground dry biomass; R/S, resistance index (ratio of GR50 of MSS or KSS and KSR populations.
b The response was fit with a three-parameter log-logistic model; fitted curves are shown in Figure 2. Values in parenthesis are ±1 SE.
c R/S values based on KSS population. R/S is significantly greater than 1 at *P<0.001.
Molecular Basis of ALS-Inhibitor Resistance
High levels of chlorsulfuron resistance in Palmer amaranth observed in our dose–response experiments indicate that the resistance may have evolved due to an alteration in the molecular target of the ALS inhibitors (ALS gene). To investigate whether mutations in the ALS gene contribute to the chlorsulfuron resistance in Palmer amaranth, the ALS gene (~2 kb) was amplified from individual plants from the KSR (survived chlorsulfuron; n=30), MSS (n=12), and KSS (n=3) populations, covering all known mutations at 8 codon positions. Only a few KSR plants showed the single-nucleotide polymorphism, resulting in an amino acid substitution of proline (CCC) to serine (TCC) at position 197 when compared with MSS or KSS (Figure 3). Interestingly, among the total 30 KSR individual plants sequenced, only 9 (30%) showed this single mutation, and the remaining 21 (70%) plants did not. A portion of the ALS gene sequence covering this mutation from only 3 each of MSS and KSS and 12 KSR plants is shown in Figure 3.
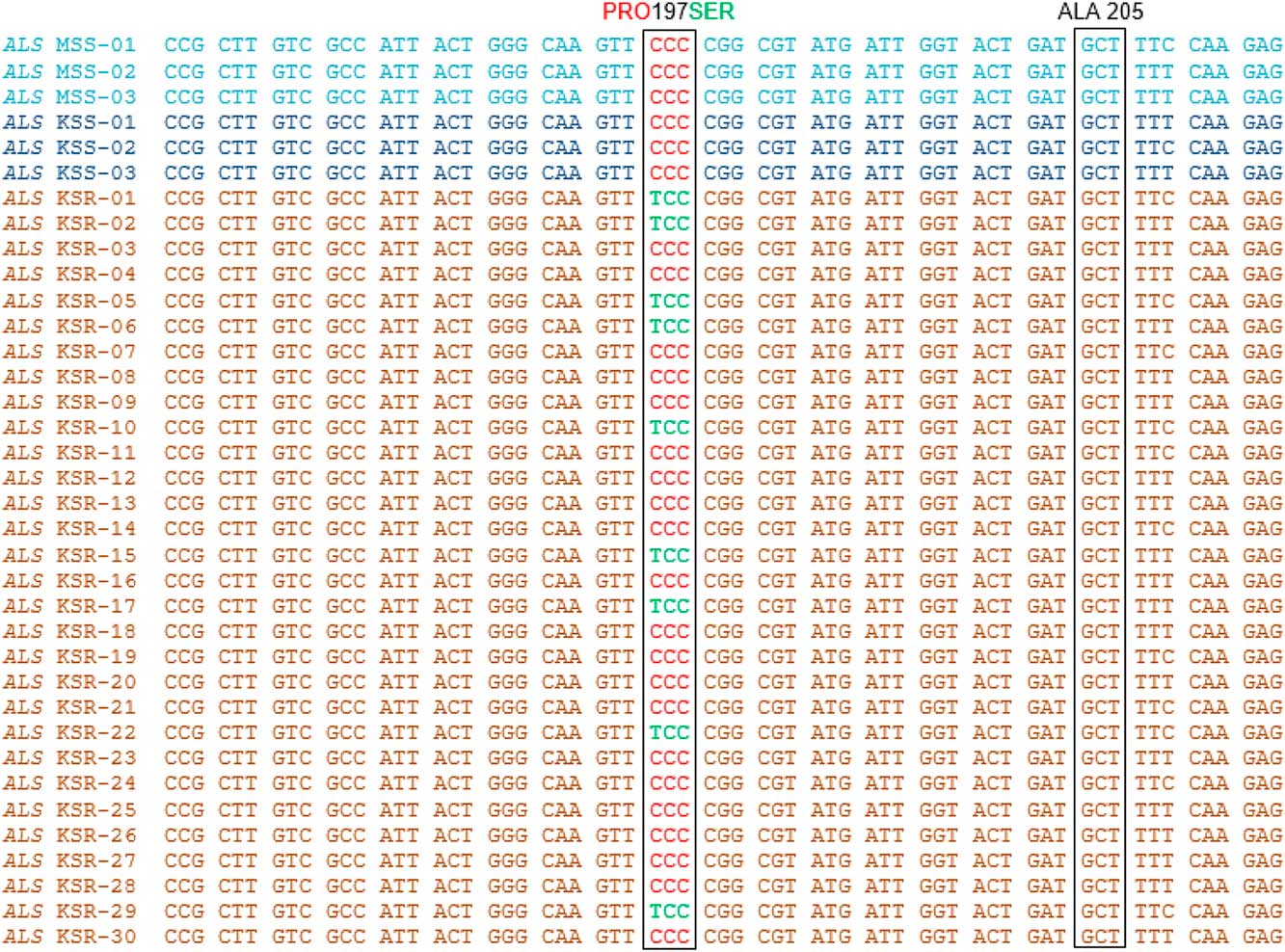
Figure 3 Nucleotide sequence alignment and analysis of a portion of ALS gene sequence from KSS, MSS, and KSR Palmer amaranth populations. Red text in the nucleotide sequence codes for proline and green text codes for serine. Nucleotide/amino acid numbering refers to the Arabidopsis thaliana ALS gene sequence.
There are a total of 11 different amino acid substitutions at proline-197 that result in resistance to most of the SU herbicides in weeds (Heap Reference Heap2017). In Amaranthus species, thus far, only two substitutions have been reported. Proline-197 is substituted by serine or leucine in prostrate pigweed (Amaranthus blitoides S. Wats.) and redroot pigweed, respectively (Sibony et al. Reference Sibony, Michel, Haas, Rubin and Hurle2001; Sibony and Rubin Reference Sibony and Rubin2003). The present report is the first describing target-site alteration (proline to serine) in Palmer amaranth. Many other Amaranthus species such as smooth pigweed and waterhemp also have evolved resistance to ALS inhibitors due to amino acid substitutions at Ala-122-Thr, Asp-376-Glu, Trp-574-Leu, or Ser-653-Thr and have shown varying levels of resistance ranging from 60- to 3,200-fold depending on the type of substitution (Patzoldt and Tranel Reference Patzoldt and Tranel2007; Whaley et al. Reference Whaley, Wilson and Westwood2006, Reference Whaley, Wilson and Westwood2007). The Asp-376-Glu substitution is known to confer a broad spectrum of resistance to all five chemical classes of ALS inhibitors (SUs, IMIs, SCTs, TPs, and PTBs) in redroot pigweed (Whaley et al. Reference Whaley, Wilson and Westwood2007). The presence of only one mutation at the proline-197 position in Palmer amaranth indicates that SUs might be the selecting agents in the evolution of resistance in the KSR population and significantly impacts the control of this weed in many SU-tolerant crops.
Nucleotide sequence analysis of the other 70% of KSR plants did not show any known amino acid substitutions on the ALS gene that confer resistance to ALS inhibitors. These results suggest the possible existence of a non–target site based resistance mechanism in our KSR Palmer amaranth population, in addition to target-site resistance to ALS inhibitors. Metabolism-based non–target site ALS-inhibitor resistance has been reported in rigid ryegrass, blackgrass (Alopecurus myosuroides Huds.), rigid brome (Bromus rigidus Roth), wild oat (Avena fatua L.), late watergrass [Echinochloa oryzicola (Vasinger) Vasinger], and wild mustard (Yu and Powles Reference Yu and Powles2014). Recently, a waterhemp population from Illinois also showed broad resistance to four chemical classes of ALS inhibitors through a metabolism-based non–target site resistance mechanism (Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015). The lack of target-site resistance in 70% of KSR individuals tested underestimates the resistance level due to target-site resistance observed in this study. Importantly, this study demonstrates for the first time the presence of both target site–based and non–target site based resistance to ALS inhibitors in a single Palmer amaranth population.
Effect of Malathion Pretreatment on Chlorsulfuron Resistance
Malathion, an organophosphate insecticide, has been used as an inhibitor of cytochrome P450 activity to study non–target site metabolism based resistance to chlorsulfuron and primsulfuron in corn and ryegrass (Christopher et al. Reference Christopher, Preston and Powles1994; Kreuz and Fonne-Pfister Reference Kreuz and Fonné-Pfister1992; Owen et al. Reference Owen, Goggin and Powles2012). Whole-plant response of KSR Palmer amaranth treated with malathion alone did not have any significant difference in the dry biomass relative to untreated control (Table 3). However, when compared with nontreated control, there was substantial difference in biomass of KSR plants treated with chlorsulfuron. In addition, the dry biomass of KSR was reduced significantly following malathion plus chlorsulfuron application relative to malathion or chlorsulfuron alone and nontreated control treatments (Table 3). The Colby analysis also showed that the expected biomass reduction achieved by malathion plus chlorsulfuron treatment was less than their respective observed reduction at 3 WAT (Table 3), indicating that malathion had a synergistic effect on chlorsulfuron. These results from treatment with malathion experiments are consistent with studies conducted in other weeds such as tall waterhemp and rigid ryegrass (Christopher et al. Reference Christopher, Preston and Powles1994; Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015). These data imply that cytochrome P450s plays an important role in the metabolism-based non–target site resistance in KSR to SU herbicides. However, it is important to separate plants with target site–based or non–target site based resistance within the KSR population to deduce accurate reduction in biomass caused by malathion. Experiments are in progress to generate a homogeneous Palmer amaranth population having no mutation to elucidate the non–target based ALS-inhibitor resistance in Palmer amaranth.
Table 3 Whole-plant response to malathion plus chlorsulfuron and chlorsulfuron alone applied to KSR Palmer amaranth (10- to 12-cm tall) when treated with malathion at 2,000 g ai ha−1.Footnote a

a Observed and expected biomass reduction of Palmer amaranth by chlorsulfuron and malathion applied alone or in combination 3 WAT. Means within columns with no common letter(s) are significantly different according to Tukey-Kramer’s pairwise comparison test at P=0.05.
b Percent biomass reduction data of nontreated control were not included in the analysis. Palmer amaranth biomass data were converted into percent biomass reduction compared with the untreated control plots using the following formula: biomass reduction (%)=[(C−B)/C]×100, where C is the biomass of the untreated control and B is the biomass collected from treated plants.
c Expected value determined by the Colby equation: E=(X+Y)−(XY/100), where E is expected biomass reduction (%) with malathion+chlorsulfuron and X and Y are observed biomass reduction (%) with malathion and chlorsulfuron, respectively.
d Significantly different from the observed value (P<0.05) as determined by t test, indicating synergism of malathion+chlorsulfuron.
Cross-resistance to ALS Inhibitors
Lack of known mutations and the malathion data provide a strong indication for the existence of a non–target site resistance mechanism to ALS inhibitors in the KSR population. Presence of a non–target site based mechanism is known to confer cross-resistance to other chemical classes of ALS inhibitors similar to some target-site mutations (e.g., Trp574Leu). We investigated the cross-resistance of KSR Palmer amaranth to thifensulfuron, imazamox, propoxycarbazone, and pyrithiobac, which belong to different families of ALS inhibitors, at field recommended rates. The results suggest that KSR Palmer amaranth was also resistant to thifensulfuron, propoxycarbazone, and pyrithiobac, but not imazamox (Table 4). The susceptibility of KSR Palmer amaranth from Stafford County to IMIs provides a viable option to control this SU-, SCT-, and PTB-resistant population in IMI-tolerant crops such as corn, wheat, canola (Brassica napus L.), rice, and common sunflower (Helianthus annuus L.) (Shaner et al. Reference Shaner, Bascomb and Smith1996; Tan et al. Reference Tan, Evans, Dahmer, Singh, Shaner, Duke and Ragsdale2005). The frequency of resistance to thifensulfuron, propoxycarabzone, or pyrithiobac in the KSR Palmer amaranth was approximately 70% to 75%, similar to chlorsulfuron resistance (Table 4). These results suggest that a non–target site mechanism, probably via cytochrome P450–mediated detoxification, may contribute resistance to SUs, SCTs, and PBTs in the KSR Palmer amaranth.
Table 4 Whole-plant response (survival data) of the KSR Palmer amaranth (10- to 12-cm tall) treated with herbicides in different chemical classes of ALS inhibitors at their field rates at 3 WAT.

a A total of 32 plants were treated and the number of plants that survived was expressed as the percent (%) survival.
ALS gene sequence analysis revealed the presence of both target-site and non–target site resistance mechanisms in KSR Palmer amaranth. Although previous research using an ALS enzyme-inhibition assay in another Palmer amaranth population indicated that resistance was due to insensitive ALS enzyme (Sprague et al. Reference Sprague, Stoller, Wax and Horak1997), our study provides evidence for target-site alteration (Pro-197-Ser) conferring resistance to sulfonylurea herbicides in ~30% of our KSR Palmer amaranth population. Nonetheless, the majority (~70%) of the KSR plants that are resistant to chlorsulfuron did not show any known mutations (Figure 3). The occurrence of both target-site and non–target site resistance mechanisms in the same population of weed species is on the rise and usually is masked by target-site resistance. The other weed species with a combination of both target site–based and non–target site based resistance to ALS inhibitors include rigid ryegrass (Christopher et al. Reference Christopher, Powles and Holtum1992), corn poppy (Papaver rhoeas L.) (Délye et al. Reference Délye, Pernin and Scarabel2011), and tall waterhemp (Guo et al. Reference Guo, Riggins, Hausman, Hager, Riechers, Davis and Tranel2015). In conclusion, evolution of target site–based and non–target site based resistance in weed species poses a serious threat to weed management, as such resistance predisposes weeds to evolve resistance to herbicides with multiple modes of action. More importantly, the dioecious nature of Palmer amaranth, which contributes to high genetic variability, combined with high seed production and efficient pollen and seed distribution (Steckel Reference Steckel2007) may help facilitate the evolution of resistance to other commonly used herbicides. Thus, management of this economically important weed is a challenge in many cropping systems. Stringent management strategies such as reducing the selection pressure of herbicides by rotating crops and herbicides and possibly tillage are warranted.
Acknowledgments
This manuscript is approved for publication as Kansas Agricultural Experiment Station Contribution no. 17-231-J.









