Impact statement
Genetic data provide useful information to guide the siting and design of conservation areas in coastal systems, such as marine protected areas. For example, reconstructing the evolutionary relationships between species through a genetic-based phylogenetic tree can inform on the presence of evolutionarily distinct species and orient the creation of marine protected areas toward sites where these species are present. Another useful application of genetic data is parentage analysis, where juveniles can be assigned to their parents and the distance between them can be used to infer the dispersal capacities of these individuals in the larval stage. These data, in turn, can be used to define the spacing between different marine protected areas, so that larvae can disperse between them while minimizing the risk of being transported to areas open to fishing. We review how applications of genetic data (including phylogenetic inference, study of intraspecific genetic variation, estimation of dispersal, and sequencing of environmental DNA) can be fruitfully used to plan networks of marine conservation areas that are more effective, meaning that they protect all facets of biodiversity in the long term. The integration of genetic data into marine spatial conservation planning can thus help reach global goals of biodiversity conservation.
Introduction
The biodiversity of coastal ecosystems is subject to a variety of human pressures, such as water pollution, overfishing, and coastal development (Andrello et al., Reference Andrello, Darling, Wenger, Suárez-Castro, Gelfand and Ahmadia2022b; Herbert-Read et al., Reference Herbert-Read, Thornton, Amon, Birchenough, Côté, Dias, Godley, Keith, McKinley, Peck, Calado, Defeo, Degraer, Johnston, Kaartokallio, Macreadie, Metaxas, Muthumbi, Obura, Paterson, Piola, Richardson, Schloss, Snelgrove, Stewart, Thompson, Watson, Worthington, Yasuhara and Sutherland2022). Marine conservation areas (MCAs), which include marine protected areas (MPAs), marine reserves, and other effective area-based conservation measures (OECMs), can partially mitigate the impacts of these pressures (Maxwell et al., Reference Maxwell, Cazalis, Dudley, Hoffmann, Rodrigues, Stolton, Visconti, Woodley, Kingston, Lewis, Maron, Strassburg, Wenger, Jonas, Venter and Watson2020; Grorud-Colvert et al., Reference Grorud-Colvert, Sullivan-Stack, Roberts, Constant, Horta e Costa, Pike, Kingston, Laffoley, Sala, Claudet, Friedlander, Gill, Lester, Day, Gonçalves, Ahmadia, Rand, Villagomez, Ban, Gurney, Spalding, Bennett, Briggs, Morgan, Moffitt, Deguignet, Pikitch, Darling, Jessen, Hameed, di Carlo, Guidetti, Harris, Torre, Kizilkaya, Agardy, Cury, Shah, Sack, Cao, Fernandez and Lubchenco2021; Gurney et al., Reference Gurney, Darling, Ahmadia, Agostini, Ban, Blythe, Claudet, Epstein, Estradivari, Himes-Cornell, Jonas, Armitage, Campbell, Cox, Friedman, Gill, Lestari, Mangubhai, McLeod, Muthiga, Naggea, Ranaivoson, Wenger, Yulianto and Jupiter2021). The Kunming–Montreal Global Biodiversity Framework of the Convention on Biological Diversity recognizes that conservation areas are pivotal for biodiversity conservation; specifically, the framework prescribes all member states to effectively conserve and manage at least 30% of coastal and marine areas “through ecologically representative, well-connected and equitably governed systems of protected areas and other effective area-based conservation measures” (CBD, 2022).
To positively impact coastal biodiversity and ecosystems, MCAs should be placed and designed following clear conservation objectives, ideally within a systematic spatial conservation planning framework (Alvarez-Romero et al., Reference Álvarez-Romero, Mills, Adams, Gurney, Pressey, Weeks, Ban, Cheok, Davies, Day, Hamel, Leslie, Magris and Storlie2018; Balbar and Metaxas, Reference Balbar and Metaxas2019). Systematic conservation planning (SCP; here used as a synonym of spatial conservation planning and spatial conservation prioritization) is a process whereby limited resources are allocated to conservation actions (such as the creation of protected areas or ecosystem restoration) that are distributed on the seascape following a criterion of optimality (Margules and Pressey, Reference Margules and Pressey2000; Margules and Sarkar, Reference Margules and Sarkar2007; Moilanen et al., Reference Moilanen, Wilson and Possingham2009b). The seascape under consideration is subdivided into planning units (PUs) of adequate size, usually dictated by the spatial scale at which the study is conducted. Each PU can be assigned to one or more conservation actions and each conservation action has a cost of implementation. In the case of MCA planning, the conservation actions are the creation of a MCA, or the zoning of a MCA into different statuses (e.g., fully protected or partially protected; Zupan et al., Reference Zupan, Fragkopoulou, Claudet, Erzini, Horta e Costa and Gonçalves2018). Costs can be quantified in monetary terms (e.g., direct costs to create and manage a marine reserve or opportunity costs representing loss of previous seascape use) or set proportional to PU size.
The criteria guiding the selection of PUs for protection are usually those of comprehensiveness, adequacy, and efficiency (Kukkala and Moilanen, Reference Kukkala and Moilanen2013). Comprehensiveness is the degree to which a set of MCAs samples the full range of biodiversity taking into account different biodiversity facets (e.g., species diversity, phylogenetic diversity, and intraspecific diversity), structure (e.g., habitat types), and functions (e.g., dispersal processes) (Wilson et al., Reference Wilson, Cabeza, Klein, Moilanen, Wilson and Possingham2009; Pollock et al., Reference Pollock, O’Connor, Mokany, Rosauer, Talluto and Thuiller2020). Adequacy means ensuring the long-term preservation of biodiversity: a common approach to address adequacy is to set conservation goals in the form of a minimum portion of species ranges covered by protected areas, but should also consider connectivity and evolutionary processes (Wilson et al., Reference Wilson, Cabeza, Klein, Moilanen, Wilson and Possingham2009; Andrello et al., Reference Andrello, D’Aloia, Dalongeville, Escalante, Guerrero, Perrier, Torres-Florez, Xuereb and Manel2022a). Efficiency means that comprehensiveness and adequacy should be fulfilled at the minimum possible cost or within a predefined budget. These criteria can be expressed in mathematical terms and formulated as a problem with explicit objectives and constraints, which can be solved using different approaches (Moilanen et al., Reference Moilanen, Possingham, Polasky, Moilanen, Wilson and Possingham2009a). The solution of the SCP problem is a list of PUs chosen for protection, which constitutes the network of MCAs for the region under study.
Molecular ecology techniques provide different types of genetic data that can inform the process of SCP in coastal marine systems (Andrello et al., Reference Andrello, D’Aloia, Dalongeville, Escalante, Guerrero, Perrier, Torres-Florez, Xuereb and Manel2022a; Jeffery et al., Reference Jeffery, Lehnert, Kess, Layton, Wringe and Stanley2022; Nielsen et al., Reference Nielsen, Hanson, Carvalho, Beger, Henriques, Kershaw and von der Heyden2022; Riginos and Beger, Reference Rabosky, Chang, Title, Cowman, Sallan, Friedman, Kaschner, Garilao, Near, Coll and Alfaro2022). First, many marine ecosystems are highly biodiverse, making it important, but challenging, to detect and map the distribution of all facets of biodiversity and resolve the evolutionary history of species. Genetic data can help resolve the phylogenetic relationships between marine species and elucidate genetic diversity within species. Second, marine environments are often challenging to sample in, leaving much biodiversity unknown. Sequencing of marine environmental DNA (eDNA; terms in italic are defined in the glossary in Table 1) is increasingly employed to complement data on the spatial distribution of species, and has the potential to lead to more comprehensive SCP solutions. Third, genetic data can inform on the existence and distribution of locally adapted populations that can be more resistant to some anthropogenic selective pressures, for example, warming waters created by climate change. Finally, planning well-connected systems of MCAs requires knowledge of dispersal of marine organisms. Genetic data can play an important role in estimating dispersal, where other approaches (such as biophysical models of larval dispersal) may fail, even if their applications to marine organisms must address the additional difficulties posed by high gene flow and large effective population sizes. With a focus on the properties of coastal marine systems, we review how four major genetic techniques (phylogenetic inference, estimation of intraspecific genetic diversity, estimation of dispersal, and environmental DNA sequencing) can be fruitfully used in SCP.
Table 1. Glossary of terms used in the text

Leveraging genetic data for marine spatial conservation planning
Phylogenetic inference
A comprehensive view of biodiversity includes multiple facets (Mace et al., Reference Mace, Gittleman and Purvis2003; Purvis et al., Reference Purvis, Gittleman and Brooks2005), one of which is the evolutionary history or evolutionary change represented by a set of taxa, usually quantified using metrics of phylogenetic diversity. Several reasons justify the consideration of evolutionary history in conservation decisions. First, conservation actions that preserve a greater amount of evolutionary history are to be favored because evolutionary history has intrinsic value like all other aspects of biodiversity. In addition, more evolutionary information is lost when a highly differentiated species from an old, species-poor clade becomes extinct than when a weakly differentiated species from a young, species-rich clade becomes extinct: this is often considered a sufficiently strong argument to prioritize phylogenetically distinct or unique species for conservation (Winter et al., Reference Winter, Devictor and Schweiger2013). Phylogenetic relationships inferred from genetic data also can help conserve biodiversity when taxonomic status is uncertain (Rosauer et al., Reference Rosauer, Byrne, Blom, Coates, Donnellan, Doughty, Keogh, Kinloch, Laver, Myers, Oliver, Potter, Rabosky, Afonso Silva, Smith and Moritz2018). From an anthropocentric point of view, conserving more evolutionary history means conserving more phenotypic diversity, which in turn can translate into enhanced benefits from ecosystem processes and potential future use of biodiversity, increased evolutionary potential, decreased extinction rates, and enhanced human experience due to a preference for nature’s variety or novelty (Tucker et al., Reference Tucker, Aze, Cadotte, Cantalapiedra, Chisholm, Díaz, Grenyer, Huang, Mazel, Pearse, Pennell, Winter and Mooers2019). A fundamental assumption underlying this point of view is that phylogenetic diversity can be a proxy for functional diversity, and is therefore linked to ecosystem functioning and services. However, the strength of the relationship between phylogenetic diversity and functional diversity varies greatly (Mazel et al., Reference Mazel, Pennell, Cadotte, Diaz, Dalla Riva, Grenyer, Leprieur, Mooers, Mouillot, Tucker and Pearse2018, Reference Mazel, Pennell, Cadotte, Diaz, Riva, Grenyer, Leprieur, Mooers, Mouillot, Tucker and Pearse2019; Owen et al., Reference Owen, Gumbs, Gray and Faith2019) and stronger evidence is needed to use phylogenetic diversity as a surrogate for functional diversity in making conservation decisions (Tucker et al., Reference Tucker, Aze, Cadotte, Cantalapiedra, Chisholm, Díaz, Grenyer, Huang, Mazel, Pearse, Pennell, Winter and Mooers2019).
High quality species-level phylogenetic trees are available for major groups of coastal organisms, including reef corals (Huang and Roy, Reference Huang and Roy2015), cartilaginous fishes (Stein et al., Reference Stein, Mull, Kuhn, Aschliman, Davidson, Joy, Smith, Dulvy and Mooers2018), ray-finned fishes (Rabosky et al., Reference Riginos, Beger, van Oppen and Aranda Lastra2018), and marine mammals (Faurby et al., Reference Faurby, Davis, Pedersen, Schowanek, Antonelli and Svenning2018; Upham et al., Reference Upham, Esselstyn and Jetz2019), while the phylogenies of other marine taxa remain less known. These unresolved phylogenies will likely benefit from the development of new sequencing technologies that have enabled “phylogenomic” approaches, in which large numbers of sequenced genes in many taxa can be used to infer phylogenetic trees (Kapli et al., Reference Kapli, Yang and Telford2020). Eventually, data from the EarthBiogenome project will allow estimating the phylogenetic tree of all eukaryotic species (Lewin et al., Reference Lewin, Richards, Lieberman Aiden, Allende, Archibald, Bálint, Barker, Baumgartner, Belov, Bertorelle, Blaxter, Cai, Caperello, Carlson, Castilla-Rubio, Chaw, Chen, Childers, Coddington, Conde, Corominas, Crandall, Crawford, DiPalma, Durbin, Ebenezer, Edwards, Fedrigo, Flicek, Formenti, Gibbs, Gilbert, Goldstein, Graves, Greely, Grigoriev, Hackett, Hall, Haussler, Helgen, Hogg, Isobe, Jakobsen, Janke, Jarvis, Johnson, Jones, Karlsson, Kersey, Kim, Kress, Kuraku, Lawniczak, Leebens-Mack, Li, Lindblad-Toh, Liu, Lopez, Marques-Bonet, Mazard, Mazet, Mazzoni, Myers, O’Neill, Paez, Park, Robinson, Roquet, Ryder, Sabir, Shaffer, Shank, Sherkow, Soltis, Tang, Tedersoo, Uliano-Silva, Wang, Wei, Wetzer, Wilson, Xu, Yang, Yoder and Zhang2022).
Gap analyses have shown that many existing systems of MCAs do not cover phylogenetic diversity adequately (Mouillot et al., Reference Mouillot, Albouy, Guilhaumon, Ben Rais Lasram, Coll, Devictor, Meynard, Pauly, Tomasini, Troussellier, Velez, Watson, Douzery and Mouquet2011; Guilhaumon et al., Reference Guilhaumon, Albouy, Claudet, Velez, Ben Rais Lasram, Tomasini, Douzery, Meynard, Mouquet, Troussellier, Araújo and Mouillot2015; May-Collado et al., Reference May-Collado, Zambrana-Torrelio, Agnarsson, Pellens and Grandcolas2016; Robuchon et al., Reference Riginos, Beger, van Oppen and Aranda Lastra2021; Mouton et al., Reference Mouton, Stephenson, Torres, Rayment, Brough, McLean, Tonkin, Albouy and Leprieur2022). For example, Mouillot et al. (Reference Mouillot, Parravicini, Bellwood, Leprieur, Huang, Cowman, Albouy, Hughes, Thuiller and Guilhaumon2016) found that the global MPA system secured only 1.7% of the tree of life for corals, and 17.6% for fishes. Moreover, spatial analyses have shown that areas with high phylogenetic diversity are also those with high taxonomic richness (Mouillot et al., Reference Mouillot, Albouy, Guilhaumon, Ben Rais Lasram, Coll, Devictor, Meynard, Pauly, Tomasini, Troussellier, Velez, Watson, Douzery and Mouquet2011; Albouy et al., Reference Albouy, Delattre, Mérigot, Meynard and Leprieur2017), but do not always overlap with areas with high functional diversity (Mouillot et al., Reference Mouillot, Albouy, Guilhaumon, Ben Rais Lasram, Coll, Devictor, Meynard, Pauly, Tomasini, Troussellier, Velez, Watson, Douzery and Mouquet2011; Mazel et al., Reference Mazel, Pennell, Cadotte, Diaz, Dalla Riva, Grenyer, Leprieur, Mooers, Mouillot, Tucker and Pearse2018; Ng et al., Reference Ng, Chisholm, Carrasco, Darling, Guilhaumon, Mooers, Tucker, Winter and Huang2022), highlighting the need to consider these biodiversity facets explicitly in SCP (Pollock et al., Reference Pollock, O’Connor, Mokany, Rosauer, Talluto and Thuiller2020). A common method to maximize phylogenetic diversity in SCP is to use the branches of the phylogenetic tree as biodiversity features (Rodrigues and Gaston, Reference Rodrigues and Gaston2002).
Finally, although we have emphasized species-level phylogenies, we note that considerable genealogical variation exists within species, so the phylogenetic approach can also be applied to integrate within-species genetic diversity into SCP (Carvalho et al., Reference Carvalho, Velo-Antón, Tarroso, Portela, Barata, Carranza, Moritz and Possingham2017).
Estimation of intraspecific genetic diversity
Intraspecific phenotypic diversity, in the form of phenotypic differentiation between individuals and populations, can be an asset allowing species to adapt to novel environmental conditions (Donelson et al., Reference Donelson, Sunday, Figueira, Gaitán-Espitia, Hobday, Johnson, Leis, Ling, Marshall, Pandolfi, Pecl, Rodgers, Booth and Munday2019). Such intraspecific phenotypic diversity emerges from the interaction between environmental variability and genetic diversity. The importance of genetic diversity has now been recognized in applied conservation (Hoban et al., Reference Hoban, Bruford, D’Urban Jackson, Lopes-Fernandes, Heuertz, Hohenlohe, Paz-Vinas, Sjögren-Gulve, Segelbacher, Vernesi, Aitken, Bertola, Bloomer, Breed, Rodríguez-Correa, Funk, Grueber, Hunter, Jaffe, Liggins, Mergeay, Moharrek, O’Brien, Ogden, Palma-Silva, Pierson, Ramakrishnan, Simo-Droissart, Tani, Waits and Laikre2020, Reference Hoban, Archer, Bertola, Bragg, Breed, Bruford, Coleman, Ekblom, Funk, Grueber, Hand, Jaffé, Jensen, Johnson, Kershaw, Liggins, MacDonald, Mergeay, Miller, Muller-Karger, O–Brien, Paz-Vinas, Potter, Razgour, Vernesi and Hunter2022). For example, the Kunming–Montreal Global Biodiversity Framework commits parties to preserve the genetic diversity of all species (CBD, 2022), including all wild species and not only crops or domestic animals as was prescribed by the Aichi targets (CBD, 2010). This objective is also motivated by mounting evidence of large declines in intraspecific genetic diversity in wild species (Leigh et al., Reference Leigh, Hendry, Vázquez-Domínguez and Friesen2019; Exposito-Alonso et al., Reference Exposito-Alonso, Booker, Czech, Gillespie, Hateley, Kyriazis, Lang, Leventhal, Nogues-Bravo, Pagowski, Ruffley, Spence, Toro Arana, Weiß and Zess2022).
In the context of SCP, intraspecific genetic diversity can be partitioned into an alpha component, measuring genetic diversity within PUs, and a beta component, measuring genetic differentiation between different PUs (Gaggiotti et al., Reference Gaggiotti, Chao, Peres-Neto, Chiu, Edwards, Fortin, Jost, Richards and Selkoe2018; Jost et al., Reference Jost, Archer, Flanagan, Gaggiotti, Hoban and Latch2018). This partitioning is relevant to understanding how different populations contribute to the genetic diversity of the species at the seascape scale, the so-called gamma diversity (Donati et al., Reference Donati, Zemp, Manel, Poirier, Claverie, Ferraton, Gaboriau, Govinden, Hagen, Ibrahim, Mouillot, Leblond, Julius, Velez, Zareer, Ziyad, Leprieur, Albouy and Pellissier2021). When gene flow is sufficiently high and effective population sizes are large, as in most marine species, PUs will be weakly differentiated and beta diversity will be close to zero, while alpha and gamma diversity will be similar (Donati et al., Reference Donati, Zemp, Manel, Poirier, Claverie, Ferraton, Gaboriau, Govinden, Hagen, Ibrahim, Mouillot, Leblond, Julius, Velez, Zareer, Ziyad, Leprieur, Albouy and Pellissier2021). In such cases, protecting a relatively small number of PUs could be sufficient for protecting a high level of gamma diversity. In some cases, though, stronger levels of genetic differentiation can persist when oceanographic features act as barriers to gene flow (Pascual et al., Reference Pascual, Rives, Schunter and Macpherson2017; Vilcot et al., Reference Vilcot, Albouy, Donati, Claverie, Julius, Manel, Pellissier and Leprieur2023), or when life history traits (e.g., lack of a pelagic larval stage) limit species’ dispersal capacity (Puritz et al., Reference Puritz, Keever, Addison, Barbosa, Byrne, Hart, Grosberg and Toonen2017). In these cases, the beta component of genetic diversity will be of significant consideration in SCP.
Different metrics and approaches can be used to measure the alpha, beta and gamma components of genetic diversity and to prioritize sites for protection. For example, Nielsen et al. (Reference Nielsen, Beger, Henriques, Selkoe and von der Heyden2017) used haplotype diversity and nucleotide diversity as metrics of alpha diversity in five coastal marine species to build an MCA system encompassing PUs with different levels of local genetic diversity. An alternative approach is to use the spatial distribution of alleles as biodiversity features and to define conservation objectives for the gamma level of genetic diversity. For example, Paz-Vinas et al. (Reference Paz-Vinas, Loot, Hermoso, Veyssière, Poulet, Grenouillet and Blanchet2018) used SCP to reach regional-level targets of representation for different alleles at microsatellite loci in six species of freshwater fishes.
A further distinction can be made between neutral genetic diversity and adaptive genetic diversity according to the effects of genetic variation on individual and population fitness (Holderegger et al., Reference Holderegger, Kamm and Gugerli2006). Partitioning of genetic diversity into neutral and adaptive components is commonly achieved using outlier tests and environmental association analyses (Hoban et al., Reference Hoban, Kelley, Lotterhos, Antolin, Bradburd, Lowry, Poss, Reed, Storfer and Whitlock2016; Manel et al., Reference Manel, Perrier, Pratlong, Abi-Rached, Paganini, Pontarotti and Aurelle2016). These techniques have suggested the existence of genetically based local adaptations to environmental conditions (e.g., water temperature, salinity, and oxygen concentration) despite extensive gene flow and lack of neutral genetic structure (e.g., Sandoval-Castillo et al., Reference Sandoval-Castillo, Robinson, Hart, Strain and Beheregaray2018; Xuereb et al., Reference Xuereb, Kimber, Curtis, Bernatchez and Fortin2018; Boulanger et al., Reference Boulanger, Benestan, Guerin, Dalongeville, Mouillot and Manel2022; Dorant et al., Reference Dorant, Laporte, Rougemont, Cayuela, Rochette and Bernatchez2022). Indeed, coastal environmental conditions are expected to change in the future and in many cases have already seen dramatic shifts, including temperature, salinity, and pH, in conjunction with local anthropogenic stressors (He and Silliman, Reference He and Silliman2019). Consequently, characterizing genetic adaptation is likely to be important for conservation in a climate change context by identifying populations that may harbor pre-adapted genetic variants and that may be able to contribute to the genetic rescue of other vulnerable populations (Bay and Palumbi, Reference Bay and Palumbi2014; Bay et al., Reference Bay, Rose, Logan and Palumbi2017; Matz et al., Reference Matz, Treml and Haller2020).
Neutral and adaptive loci, and sets of loci associated with different environmental variables, can have markedly different spatial distributions (Barbosa et al., Reference Barbosa, Mestre, White, Paupério, Alves and Searle2018; Sandoval-Castillo et al., Reference Sandoval-Castillo, Robinson, Hart, Strain and Beheregaray2018); therefore, spatial protection priorities identified through SCP can vary (Hanson et al., Reference Hanson, Marques, Veríssimo, Camacho-Sanchez, Velo-Antón, Martínez-Solano and Carvalho2020). For example, Xuereb et al. (Reference Xuereb, D’Aloia, Andrello, Bernatchez and Fortin2021a) identified different sets of priority PUs for protection of genetic diversity for the sea cucumber Parastichopus californicus in coastal British Columbia (Canada) depending on whether they considered neutral or putatively adaptive loci. The choice of metrics to measure adaptive genetic diversity also led to different spatial conservation priorities (Xuereb et al., Reference Xuereb, D’Aloia, Andrello, Bernatchez and Fortin2021a).
Estimation of dispersal
The importance of connectivity for the conservation and management of marine species has long been recognized and prescribed as an important criterion for siting, sizing, and designing MCAs (Palumbi, Reference Palumbi2003; Alvarez-Romero et al., Reference Álvarez-Romero, Mills, Adams, Gurney, Pressey, Weeks, Ban, Cheok, Davies, Day, Hamel, Leslie, Magris and Storlie2018; Balbar and Metaxas, Reference Balbar and Metaxas2019). Indeed, determining whether the set of prioritized MCAs truly represents a connected “network” depends on the strength of connectivity between them. This criterion is emphasized in both the Aichi targets and the Kunming–Montreal Global Biodiversity Framework of the Convention on Biological Diversity, which requires that MCAs form a “well-connected” system (CBD, 2010, 2022).
Networks of MCAs should satisfy different connectivity-related objectives (Beger et al., Reference Beger, Metaxas, Balbar, McGowan, Daigle, Kuempel, Treml and Possingham2022; Riginos and Beger, Reference Rabosky, Chang, Title, Cowman, Sallan, Friedman, Kaschner, Garilao, Near, Coll and Alfaro2022). In particular, MCAs should be strategically designed, placed and spaced to protect foraging movements in the home range of species and ontogenetic migrations between habitats for different life cycle stages (Grüss et al., Reference Grüss, Kaplan, Guenette, Roberts and Botsford2011; D’Aloia et al., Reference D’Aloia, Daigle, Côté, Curtis, Guichard and Fortin2017). Furthermore, MCA networks should ensure demographic connectivity, including replenishment of “sink” populations and recolonization of empty habitat patches (Almany et al., Reference Almany, Berumen, Thorrold, Planes and Jones2007; Saenz-Agudelo et al., Reference Saenz-Agudelo, Jones, Thorrold and Planes2011; Harrison et al., Reference Harrison, Bode, Williamson, Berumen and Jones2020), and genetic connectivity, particularly the spread of advantageous genetic variants allowing for genetic rescue of imperiled populations (Webster et al., Reference Webster, Colton, Darling, Armstrong, Pinsky, Knowlton and Schindler2017; Bell et al., Reference Bell, Robinson, Funk, Fitzpatrick, Allendorf, Tallmon and Whiteley2019). Finally, connectivity allows marine reserves to support fisheries outside their borders through adult spillover and larval export (Andrello et al., Reference Andrello, Guilhaumon, Albouy, Parravicini, Scholtens, Verley, Barange, Sumaila, Manel and Mouillot2017; Di Lorenzo et al., Reference Di Lorenzo, Guidetti, Di Franco, Calò and Claudet2020; Medoff et al., Reference Medoff, Lynham and Raynor2022). Various operational approaches have been developed to integrate these aspects of connectivity into SCP (Daigle et al., Reference Daigle, Metaxas, Balbar, McGowan, Treml, Kuempel, Possingham and Beger2020; Beger et al., Reference Beger, Metaxas, Balbar, McGowan, Daigle, Kuempel, Treml and Possingham2022). These methods use node-based or link-based connectivity metrics (Table 2): PUs are represented as nodes (vertices) of a network, and connections (in the form of larval dispersal probabilities, gene flow or spatial distances between pairs of PUs) are the links (edges) between the nodes (Xuereb et al., Reference Xuereb, D’Aloia, Daigle, Andrello, Dalongeville, Manel, Mouillot, Guichard, Côté, Curtis, Bernatchez, Fortin, Oleksiak and Rajora2020).
Table 2. Methods to integrate connectivity into spatial conservation planning (SCP)
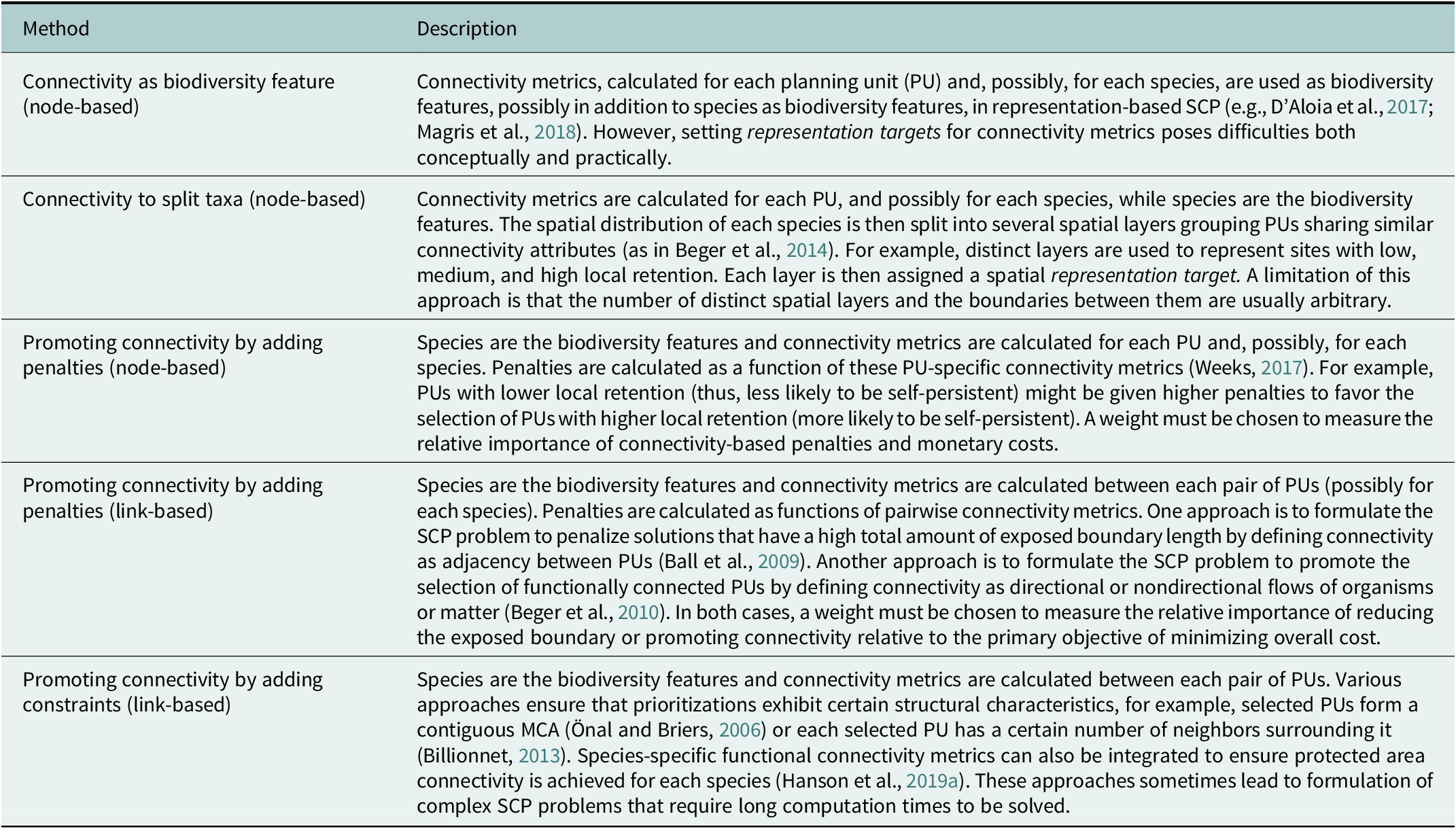
Genetic data can be used to estimate aspects of connectivity through the effects of dispersal (an individual-level process linked to connectivity; Baguette et al., Reference Baguette, Blanchet, Legrand, Stevens and Turlure2013) on population genetics. More precisely, genetic data are used to estimate both noneffective dispersal (where the dispersing agent moves into another habitat regardless of whether it successfully reproduces and transmits its genes) and effective dispersal (when the disperser successfully transmits its genes) (Cayuela et al., Reference Cayuela, Rougemont, Prunier, Moore, Clobert, Besnard and Bernatchez2018).
Noneffective dispersal can be estimated by individual-level genetic approaches such as population assignment tests and parentage analysis (Broquet and Petit, Reference Broquet and Petit2009; Cayuela et al., Reference Cayuela, Rougemont, Prunier, Moore, Clobert, Besnard and Bernatchez2018). Assignment tests can inform on dispersal when individuals are confidently assigned to genetically differentiated groups or populations (Manel et al., Reference Manel, Gaggiotti and Waples2005; Christie et al., Reference Christie, Meirmans, Gaggiotti, Toonen and White2017). Given these requirements, assignment tests are impractical for many marine species with large population sizes, high mobility, and weak population structure; however, they have been successfully applied to detect long-distance dispersal in a coral reef fish (D’Aloia et al., Reference D’Aloia, Bogdanowicz, Andrés and Buston2022). In contrast, parentage analysis has been repeatedly used to estimate dispersal in coastal fishes, including studies on dispersal between MPAs and larval supply from marine reserves to fished areas (e.g., Harrison et al., Reference Harrison, Williamson, Evans, Almany, Thorrold, Russ, Feldheim, van Herwerden, Planes, Srinivasan, Berumen and Jones2012; Almany et al., Reference Almany, Hamilton, Bode, Matawai, Potuku, Saenz-Agudelo, Planes, Berumen, Rhodes, Thorrold, Russ and Jones2013; Baetscher et al., Reference Baetscher, Anderson, Gilbert-Horvath, Malone, Saarman, Carr and Garza2019). With appropriate marker panels, these methods are highly accurate, but usually only assign a small percentage of sampled individuals (Christie et al., Reference Christie, Meirmans, Gaggiotti, Toonen and White2017): therefore, they require extensive sampling of possible offspring and parents, which can limit their applications to relatively small populations and/or study areas. To date, parentage studies have also been taxonomically biased toward fishes, but there is promise in their application to invertebrates, such as corals (Dubé et al., Reference Dubé, Boissin, Mercière and Planes2020).
Effective dispersal can be estimated through simple relationships between migration rates and indices of genetic differentiation between populations (e.g., F ST), but the assumptions of these relationships (such as equal size across populations; Whitlock and McCauley, Reference Whitlock and McCauley1999) are rarely met in natural populations, making this approach unreliable. Patterns of isolation-by-distance (IBD), where genetic similarity between individuals or populations decreases with spatial distance, can be used to estimate dispersal distances under the assumption of migration–drift equilibrium and knowledge of effective population density (e.g., Puebla et al., Reference Puebla, Bermingham and McMillan2012; Benestan et al., Reference Benestan, Fietz, Loiseau, Guerin, Trofimenko, Rühs, Schmidt, Rath, Biastoch, Pérez-Ruzafa, Baixauli, Forcada, Arcas, Lenfant, Mallol, Goñi, Velez, Höppner, Kininmonth, Mouillot, Puebla and Manel2021). When applied to two coral reef fishes with limited dispersal potential, IBD approaches provided estimates of effective dispersal distances that were very similar to estimates of noneffective dispersal obtained through parentage analysis (Pinsky et al., Reference Pinsky, Saenz-Agudelo, Salles, Almany, Bode, Berumen, Andréfouët, Thorrold, Jones and Planes2017; Naaykens and D’Aloia, Reference Naaykens and D’Aloia2022). Future work on other taxa will reveal whether these relationships hold for more dispersive species.
To date, only one marine SCP study has incorporated genetically derived estimates of dispersal rates (Beger et al., Reference Beger, Selkoe, Treml, Barber, von der Heyden, Crandall, Toonen and Riginos2014). However, in recent years, genetic studies of marine dispersal have increased in number (Xuereb et al., Reference Xuereb, D’Aloia, Daigle, Andrello, Dalongeville, Manel, Mouillot, Guichard, Côté, Curtis, Bernatchez, Fortin, Oleksiak and Rajora2020), widened in scope (from single species to multi species (Benestan et al., Reference Benestan, Fietz, Loiseau, Guerin, Trofimenko, Rühs, Schmidt, Rath, Biastoch, Pérez-Ruzafa, Baixauli, Forcada, Arcas, Lenfant, Mallol, Goñi, Velez, Höppner, Kininmonth, Mouillot, Puebla and Manel2021); from a single year to multiple years (Catalano et al., Reference Catalano, Dedrick, Stuart, Puritz, Montes and Pinsky2021); and from the detection of few dispersal events to the estimation of full connectivity matrices (Dedrick et al., Reference Dedrick, Catalano, Stuart, White, Montes and Pinsky2021)), and seemingly improved in accuracy, as demonstrated by the concordance of dispersal estimates obtained with different approaches (D’Aloia et al., Reference D’Aloia, Bogdanowicz, Francis, Majoris, Harrison and Buston2015, Reference D’Aloia, Xuereb, Fortin, Bogdanowicz and Buston2018, Reference D’Aloia, Bogdanowicz, Andrés and Buston2022; Pinsky et al., Reference Pinsky, Saenz-Agudelo, Salles, Almany, Bode, Berumen, Andréfouët, Thorrold, Jones and Planes2017; Bode et al., Reference Bode, Leis, Mason, Williamson, Harrison, Choukroun and Jones2019). Moreover, there is potential to gain additional dispersal data from other genetic approaches such as clinal analyses (Gagnaire et al., Reference Gagnaire, Broquet, Aurelle, Viard, Souissi, Bonhomme, Arnaud-Haond and Bierne2015; Van Wyngaarden et al., Reference van Wyngaarden, Snelgrove, DiBacco, Hamilton, Rodríguez-Ezpeleta, Jeffery, Stanley and Bradbury2017), spatial analyses of close kin (Rueger et al., Reference Rueger, Harrison, Buston, Gardiner, Berumen and Jones2020; Benestan et al., Reference Benestan, Fietz, Loiseau, Guerin, Trofimenko, Rühs, Schmidt, Rath, Biastoch, Pérez-Ruzafa, Baixauli, Forcada, Arcas, Lenfant, Mallol, Goñi, Velez, Höppner, Kininmonth, Mouillot, Puebla and Manel2021; Jasper et al., Reference Jasper, Hoffmann and Schmidt2022), and machine learning (e.g., Smith et al., Reference Smith, Tittes, Ralph and Kern2023). If these trends continue, more datasets on marine dispersal rates and distances will become available for SCP applications.
Environmental DNA sequencing
Environmental DNA metabarcoding is a recently developed method to detect the presence of species and/or molecular operational taxonomic units (MOTUs) from DNA fragments released by organisms into their environment (Taberlet et al., Reference Taberlet, Bonin, Zinger and Coissac2018). A genetic marker (metabarcode) is chosen to target a taxonomic group (e.g., eukaryotes and teleosts), identify MOTUs, and assign MOTUs to known species if the species’ metabarcode is sequenced (Miya, Reference Miya2022). eDNA metabarcoding is recognized to outperform traditional techniques for detecting elusive species such as cryptobenthic fishes (Boulanger et al., Reference Boulanger, Loiseau, Valentini, Arnal, Boissery, Dejean, Deter, Guellati, Holon, Juhel, Lenfant, Manel and Mouillot2021; Mathon et al., Reference Mathon, Marques, Mouillot, Albouy, Andrello, Baletaud, Borrero-Pérez, Dejean, Edgar, Grondin, Guerin, Hocdé, Juhel, Kadarusman, Maire, Mariani, McLean, Polanco F., Pouyaud, Stuart-Smith, Sugeha, Valentini, Vigliola, Vimono, Pellissier and Manel2022) and rare species such as nonindigenous (Comtet et al., Reference Comtet, Sandionigi, Viard and Casiraghi2015; Duarte et al., Reference Duarte, Vieira, Lavrador and Costa2021) or threatened species (Weltz et al., Reference Weltz, Lyle, Ovenden, Morgan, Moreno and Semmens2017; Juhel et al., Reference Juhel, Marques, Utama, Vimono, Sugeha, Kadarusman, Cochet, Dejean, Hoey, Mouillot, Hocdé and Pouyaud2022). For example, eDNA metabarcoding was able to detect 44% more shark species than underwater visual censuses and baited remote underwater video station survey methods, even with a much lower sampling effort (Boussarie et al., Reference Boussarie, Bakker, Wangensteen, Mariani, Bonnin, Juhel, Kiszka, Kulbicki, Manel, Robbins, Vigliola and Mouillot2018).
As SCP requires extensive spatial occurrence data, eDNA is emerging as the method of choice to complete existing occurrence data at large spatial scales (Bani et al., Reference Bani, de Brauwer, Creer, Dumbrell, Limmon, Jompa, von der Heyden, Beger, Dumbrell, Turner and Fayle2020; Table 3). To this end, eDNA metabarcoding has recently been used to improve species distribution models of lake fishes (Pukk et al., Reference Pukk, Kanefsky, Heathman, Weise, Nathan, Herbst, Sard, Scribner and Robinson2021), deep sea fishes (McClenaghan et al., Reference McClenaghan, Compson and Hajibabaei2020), and tropical coral reefs (Jaquier et al., Reference Jaquier, Albouy, Bach, Waldock, Marques, Maire, Juhel, Andrello, Valentini, Manel, Dejean, Mouillot and Pellissierunder review). eDNA metabarcoding is also a good method to complement distributional data for multiple biodiversity facets, as shown by the similarities of phylogenetic and functional diversity measurements obtained through eDNA metabarcoding to those obtained through underwater videos (Marques et al., Reference Marques, Castagné, Polanco Fernández, Borrero-Pérez, Hocdé, Guérin, Juhel, Velez, Loiseau, Letessier, Bessudo, Valentini, Dejean, Mouillot, Pellissier and Villéger2021).
Table 3. Potential use of information gained from eDNA in spatial conservation planning (SCP), and future developments
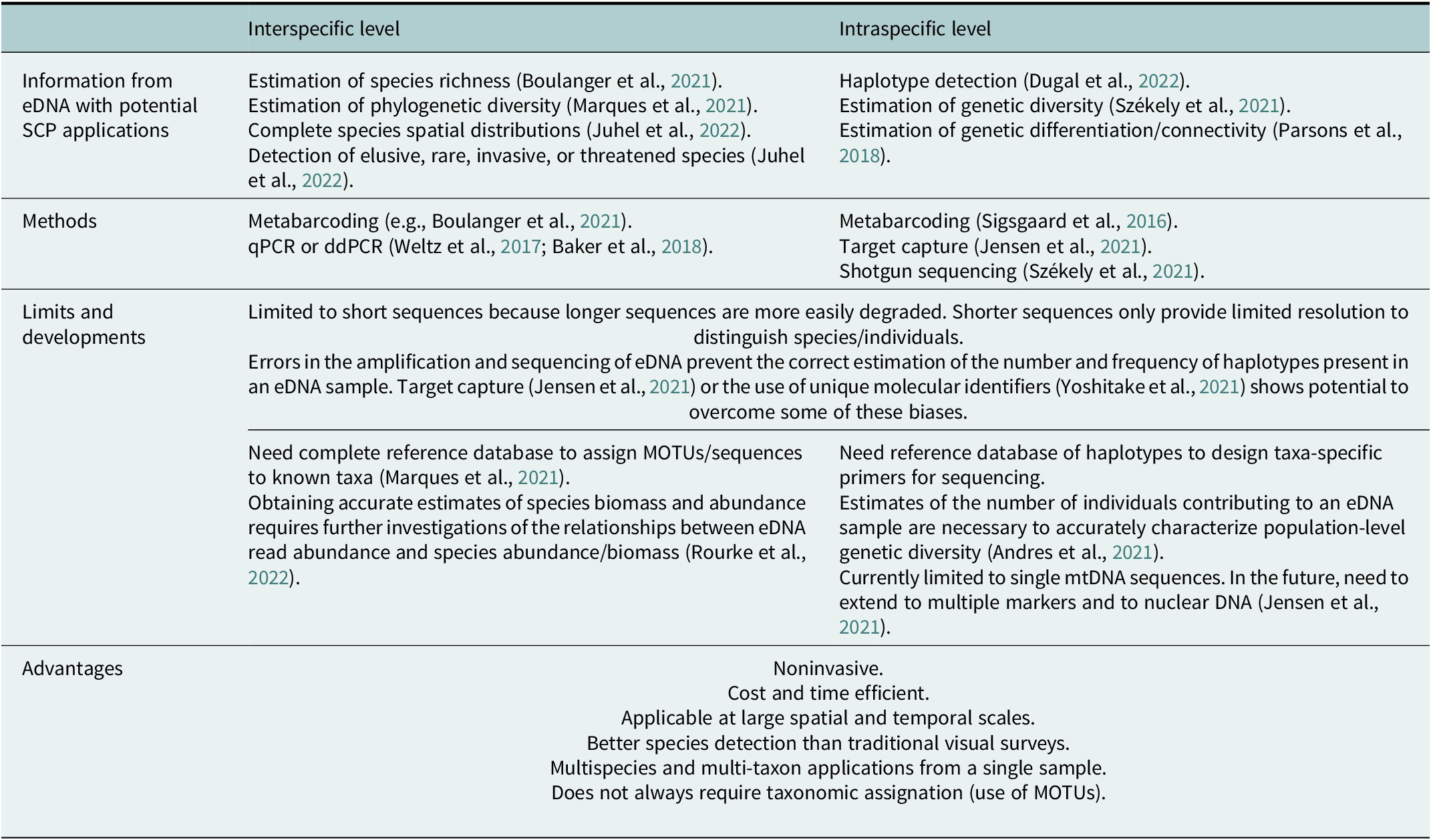
eDNA has also started to be used to study intraspecific genetic diversity (Sigsgaard et al., Reference Sigsgaard, Jensen, Winkelmann, Møller, Hansen and Thomsen2020; Table 3). While intraspecific applications of eDNA are currently limited by the nature, number, and length of markers used (see Table S1 in the Supplementary Material), they open important perspectives to the use of eDNA for estimating intraspecific genetic diversity and connectivity when tissue sampling is problematic, as for mobile, cryptobenthic or threatened species (Dugal et al., Reference Dugal, Thomas, Jensen, Sigsgaard, Simpson, Jarman, Thomsen and Meekan2022). The first intraspecific applications of eDNA were based on the metabarcoding of a single mitochondrial sequence (Sigsgaard et al., Reference Sigsgaard, Nielsen, Bach, Lorenzen, Robinson, Knudsen, Pedersen, Jaidah, Orlando, Willerslev, Møller and Thomsen2016; Macé et al., Reference Macé, Hocdé, Marques, Guerin, Valentini, Arnal, Pellissier and Manel2022). The challenge is now to extend the use of eDNA to multiple markers, including nuclear markers, to obtain more accurate estimates of intraspecific genetic diversity than with one single longer sequence. Andres et al. (Reference Andres, Sethi, Lodge and Andrés2021) developed nuclear microsatellites from eDNA and applied it to the estimation of unique genetic contributors in an experimental mesocosm of Neogobius melanostomus, showing the potential of this technique to estimate population size. The next step toward obtaining finer spatial genetic structure estimates with eDNA is to get nuclear SNP data. Target capture is thus starting to be developed on eDNA for this purpose. In one recent example, Jensen et al. (Reference Jensen, Sigsgaard, Liu, Manica, Bach, Hansen, Møller and Thomsen2021) applied target capture of nuclear markers for whale sharks Rhincodon typus on eDNA samples, but they obtained low read coverage of the targeted nuclear regions, and sequences were confounded by the highly abundant mackerel tuna Euthynnus affinis.
Successful applications to study interspecific and intraspecific diversity show that analysis of eDNA provides a potentially powerful tool to overcome the lack of spatial biodiversity data for marine SCP (Bani et al., Reference Bani, de Brauwer, Creer, Dumbrell, Limmon, Jompa, von der Heyden, Beger, Dumbrell, Turner and Fayle2020). To the best of our knowledge, only one study has used eDNA metabarcoding in a SCP framework (Mathon et al., Reference Mathon, Baletaud, Lebourges-Dhaussy, Lecellier, Menkes, Bachelier, Bonneville, Dejean, Dumas, Fiat, Grelet, Habasque, Manel, Mannocci, Mouillot, Peran, Roudaut, Sidobre, Varillon and Vigliolaunpublished results): the authors combined marine fish occurrence data from acoustic, video and eDNA metabarcoding to prioritize conservation units in a three-dimensional space across 15 seamounts and deep island slopes in the Coral Sea. eDNA metabarcoding identified almost twice as many families as baited remote underwater video stations, and 596 MOTUs versus 190 species (Mathon et al., Reference Mathon, Baletaud, Lebourges-Dhaussy, Lecellier, Menkes, Bachelier, Bonneville, Dejean, Dumas, Fiat, Grelet, Habasque, Manel, Mannocci, Mouillot, Peran, Roudaut, Sidobre, Varillon and Vigliolaunpublished results).
Conclusions and future perspectives
The studies reviewed here show that genetic data can help meet the overarching goals of comprehensiveness, adequacy, and efficiency that inspire SCP (Nielsen et al., Reference Nielsen, Hanson, Carvalho, Beger, Henriques, Kershaw and von der Heyden2022). Inclusion of phylogenetic diversity and intraspecific genetic diversity in addition to taxonomic diversity can increase the comprehensiveness of systems of MCAs, while the characterization of taxonomic diversity, itself, will benefit from the rapid generation of complementary information from eDNA techniques. The integration of connectivity and adaptive genetic diversity can help create networks of MCAs that better satisfy the adequacy criterion, because they ensure greater long-term persistence of biodiversity. In terms of economic efficiency, genetic data has the potential to both generate benefits and incur additional costs. For example, using genetic data to promote connectivity between MCAs and fished areas can increase the economic efficiency of MCAs thanks to the economic benefits that fisheries derive from increased catches. More generally, however, the integration of new facets and processes may increase the total area required to satisfy additional conservation targets and thus total conservation costs.
There are still few examples of the integration of genetic data in marine SCP, especially with regards to intraspecific genetic data (Table 4). This is mainly due to the low number of species for which spatial genetic data are available. While molecular ecology studies are being carried out on an increasing set of species, it will be important to assess whether environmental variables that are easier to obtain can be used as proxies for intraspecific genetic diversity. The few studies testing such hypotheses have yielded mixed results (Hanson et al., Reference Hanson, Rhodes, Riginos and Fuller2017, Reference Hanson, Veríssimo, Velo-Antón, Marques, Camacho-Sanchez, Martínez-Solano, Gonçalves, Sequeira, Possingham and Carvalho2021; Manel et al., Reference Manel, Guerin, Mouillot, Blanchet, Velez, Albouy and Pellissier2020) and predictive models relating environmental variables to genetic distance using isolation-by-resistance models (e.g., Boussarie et al., Reference Boussarie, Momigliano, Robbins, Bonnin, Cornu, Fauvelot, Kiszka, Manel, Mouillot and Vigliola2022) remain rare in seascape genetics and seldom tested in an SCP setting (Hanson et al., Reference Hanson, Fuller and Rhodes2019a). eDNA represents a promising alternative, as readily available material could be used to obtain information on intraspecific genetic diversity and connectivity for multiple species from a single sample.
Table 4. List of published spatial conservation planning (SCP) studies for marine and coastal habitats integrating genetic data
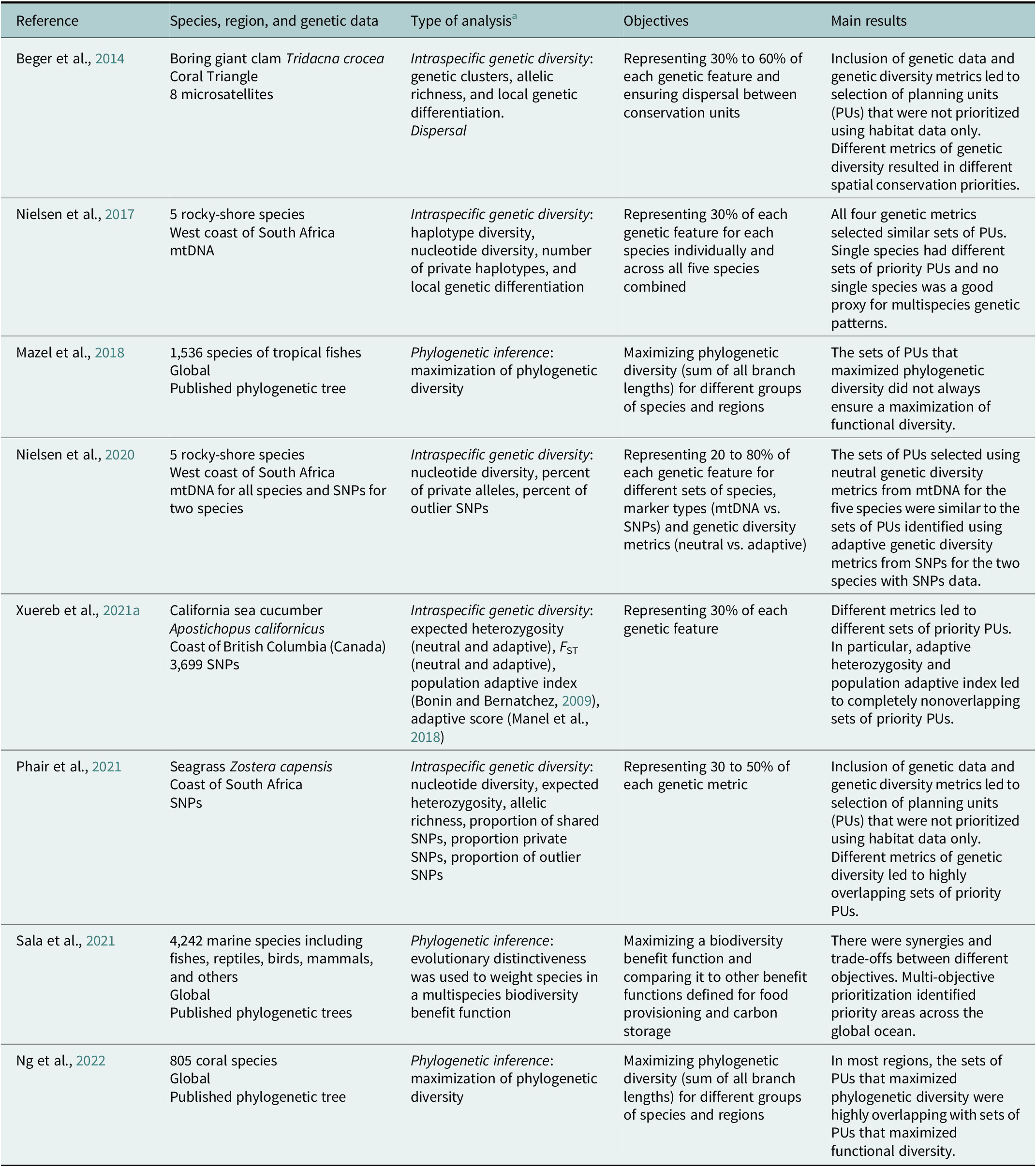
a This column indicates which of the four applications of genetic data presented in the main text (phylogenetic inference, intraspecific genetic diversity, dispersal, or eDNA) was used in the study. In the case of intraspecific genetic diversity, the genetic metrics are indicated. Note that there is no published SCP study integrating eDNA data.
In cases where species-level genetic data already exist, it is important to account for uncertainty when integrating these data into SCP. In particular, uncertainty can arise from (i) genotyping errors (Pompanon et al., Reference Pompanon, Bonin, Bellemain and Taberlet2005), (ii) information content of different molecular markers (D’Aloia et al., Reference D’Aloia, Andrés, Bogdanowicz, McCune, Harrison and Buston2020), (iii) identification of putatively adaptive and neutral loci (de Mita et al., Reference de Mita, Thuillet, Gay, Ahmadi, Manel, Ronfort and Vigouroux2013; Dalongeville et al., Reference Dalongeville, Benestan, Mouillot, Lobreaux and Manel2018), (iv) estimation of seascape genetic parameters from samples of finite size, obtained from a portion of a species’ range (parameter uncertainty; Balkenhol and Fortin, Reference Balkenhol, Fortin, Balkenhol, Cushman, Storfer and Waits2015; Foster et al., Reference Foster, Feutry, Grewe and Davies2021), (v) differences between statistical approaches used to infer seascape genetic parameters and spatial planning algorithms (model uncertainty; Hanson et al., Reference Hanson, Schuster, Strimas-Mackey and Bennett2019b), (vi) prediction of seascape genetic parameters in unsampled sites (Manel and Holderegger, Reference Manel and Holderegger2013), and (vii) availability of only a small number of species with genetic data that are used as a representative surrogate for the genetic biodiversity of all the species present in a region (Nielsen et al., Reference Nielsen, Beger, Henriques and von der Heyden2020). Recent studies have begun to integrate some of these aspects of uncertainty into genetically informed SCP (Nielsen et al., Reference Nielsen, Beger, Henriques and von der Heyden2020, Reference Nielsen, Hanson, Carvalho, Beger, Henriques, Kershaw and von der Heyden2022; Xuereb et al., Reference Xuereb, D’Aloia, Andrello, Bernatchez and Fortin2021a). Moreover, forward-in-time simulations are promising approaches for predicting spatial genetic patterns relevant for SCP and could be used to test the potential impact of conservation actions under complex eco-evolutionary scenarios (Xuereb et al., Reference Xuereb, Rougemont, Tiffin, Xue and Phifer-Rixey2021b), although there remains considerable uncertainty in the estimation of genetic and demographic parameters required for such simulations for the majority of marine species.
In spite of their low number, the published SCP studies in marine and coastal systems show that genetic data provide information that cannot be gained without them. For example, in some taxonomic groups, phylogenetic distances between species are only partially congruent with distances in functional traits, showing that genetic data are necessary to capture and prioritize the evolutionary relationships between species (Mazel et al., Reference Mazel, Pennell, Cadotte, Diaz, Dalla Riva, Grenyer, Leprieur, Mooers, Mouillot, Tucker and Pearse2018). Another important review of marine species showed that genetic-based estimates of larval dispersal can be an order of magnitude smaller than those obtained using biophysical models (Manel et al., Reference Manel, Loiseau, Andrello, Fietz, Goñi, Forcada, Lenfant, Kininmonth, Marcos, Marques, Mallol, Pérez-Ruzafa, Breusing, Puebla and Mouillot2019). Such discrepancies have direct consequences for setting optimal distances between MCAs. More generally, expanding the taxonomic, phylogenetic, geographical, and temporal scope of seascape genetic studies will confirm or refute the generality of these patterns and their consequences on the optimal design of effective MCA networks to protect coastal biodiversity. Given the ongoing increases in molecular data collection across diverse marine taxa and habitats, we anticipate that genetic and genomic data will play an increasingly prominent role in coastal SCP.
Open peer review
To view the open peer review materials for this article, please visit http://doi.org/10.1017/cft.2023.16.
Supplementary material
The supplementary material for this article can be found at http://doi.org/10.1017/cft.2023.16.
Data availability statement
Data availability is not applicable to this article as no new data were created or analyzed in this study.
Acknowledgments
We thank Richard Schuster, two other anonymous reviewers, and the senior editor Martin Le Tissier for their helpful comments. We also thank the editor Alicia Acosta and the publisher and senior scientific editor Jessica Jones for encouraging us to write this review.
Author contribution
Conceptualization: M.A.; Writing – original draft, and review and editing: M.A., S.M., M.V., A.X., and C.D.
Financial support
This work was supported by Agence Nationale de la Recherche (Grant No. ANR-18-CE02–0016 SEAMOUNTS).
Competing interest
The authors declare no competing interest.







Comments
Dear Editor,
Thanks for your invitation to write a review on the “Benefits of genetic data for spatial conservation planning in coastal habitats” for Coastal futures. Please find the manuscript attached.
In this manuscript, we illustrate how genetic data can be fruitfully used to inform spatial planning of conservation actions for coastal systems. We focused our attention on four applications: phylogenetic inference, estimation of intraspecific genetic diversity, estimation of connectivity and dispersal, and sequencing of environmental DNA. The content of our manuscript offers an original, up-to-date and detailed vision relative to three recent reviews on similar subject: namely those written by our group (Andrello et al 2022; DOI: 10.1016/j.tree.2022.03.003), by Nielsen et al 2022 (DOI: 10.1016/j.tree.2022.09.006) and by Jeffery et al 2022 (DOI: 10.3389/fgene.2022.886494). Relative to the first two papers, we explore in deeper details the challenges posed by marine systems to spatial conservation planning and use of genetic data to obtain relevant information. Relative to the third paper, we explore in more detail the use of genetic data to infer connectivity, dispersal and phylogenies. Finally, our manuscript provides a systematic review on the use of eDNA in spatial conservation planning and an updated review on the use of eDNA to study intraspecific genetic diversity.
To review this topic, I put together an international team of scientists with complementary expertise on the subject. Namely, Prof. Stéphanie Manel has a long standing expertise on seascape genetics, Ms Maurine Vilcot applies of environmental DNA for studying marine biodiversity, Dr. Amanda Xuereb has expertise on the use of genetic data to identify genetic adaptation in marine organisms and prof. Cassidy D’Aloia is an expert on the use of genetic data to infer dispersal of marine organisms.
We hope that the editorial team will like our manuscript, and look forward to your decision.
Yours faithfully,
Marco Andrello, on behalf of all authors