Introduction
Diplostomidae Poirier, 1886, is a large and globally distributed family of digeneans whose adults are found in the intestines of birds and mammals (Niewiadomska, Reference Niewiadomska, Gibson, Jones and Bray2002; Heneberg et al., Reference Heneberg, Sitko and Těšínský2020). Among diplostomids, the genus Posthodiplostomum Dubois, Reference Dubois1936, has been investigated in numerous studies related to their taxonomy, ecology, host–parasite relationships and pathogenicity (e.g. Dubois, Reference Dubois1970; Niewiadomska, Reference Niewiadomska, Gibson, Jones and Bray2002; López-Hernández et al., Reference López-Hernández, Locke, De Melo, Rabelo and Pinto2018; Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021). A recently published study on the diversity of the subfamily former Crassiphialinae Sudarikov, 1960, through molecular data proposed the synonymy of the genera Ornithodiplostomum Dubois, Reference Dubois1936 and Mesoophorodiplostomum Dubois, Reference Dubois1936 with Posthodiplostomum (Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021). According to this new taxonomic reorganization, the genus Posthodiplostomum currently contains 35 species; most species in the genus, as adults, are parasites of fish-eating birds of the family Ardeidae Leach (Dubois, Reference Dubois1970; Niewiadomska, Reference Niewiadomska, Gibson, Jones and Bray2002; López-Hernández et al., Reference López-Hernández, Locke, De Melo, Rabelo and Pinto2018; Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021). The database of DNA sequences from Posthodiplostomum has increased steadily in recent years with the availability of sequence data from global sources, expanding our knowledge of species diversity, classification and biogeography. Nevertheless, assembling a comprehensive molecular database has been challenging because various authors have sequenced different nuclear regions, e.g. the D2–D3 or D1–D3 domains of the large (28S) or small subunit (18S), the transcribed spacers (ITS1–5.8S–ITS2), and different regions of the mitochondrial gene as the first region the 5′ beginning (typical barcoding region) or second region the 3′end of cytochrome oxidase (cox1) (see Locke et al., Reference Locke, McLaughlin and Marcogliese2010; Nguyễn et al., Reference Nguyễn, Li, Makouloutou, Jimenez and Sato2012; Kvach et al., Reference Kvach, Jurajda, Bryjová, Trichkova, Ribeiro, Přikrylová and Ondračková2017; Stoyanov et al., Reference Stoyanov, Georgieva, Pankov, Kudlai, Kostadinova and Georgiev2017; Boone et al., Reference Boone, Laursen, Colombo, Meiners, Romani and Keeney2018; López-Hernández et al., Reference López-Hernández, Locke, De Melo, Rabelo and Pinto2018; Hoogendoorn et al., Reference Hoogendoorn, Smit and Kudlai2019; Sokolov and Gordeev, Reference Sokolov and Gordeev2020; Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021; Duan et al., Reference Duan, Al-Jubury, Kania and Buchmann2021; Pernett et al., Reference Pernett, Brant and Locke2022; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022).
The only species of Posthodiplostomum known to parasitize fish and fish-eating birds across Mexico was P. minimum (McCallum, 1921) Dubois, Reference Dubois1936 (Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García-Prieto and Mendoza-Garfías2007). However, extensive sampling of metacercariae and adults of Posthodiplostomum and the use of molecular tools allowed us to uncover a large species diversity in the genus. For example, Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022 identified 6 genetic lineages in what was once considered a single species. The metacercariae of P. minimum have been reported from 109 fish species and adults from 7 species of fish-eating birds (Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García-Prieto and Mendoza-Garfías2007, Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022). However, no molecular data are available for adults, preventing the establishment of a link between larval forms and adults and the recognition of candidate species instead of only genetic lineages (Locke et al., Reference Locke, McLaughlin and Marcogliese2010; López-Hernández et al., Reference López-Hernández, Locke, De Melo, Rabelo and Pinto2018; Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022).
Here, we filled out the knowledge gap concerning the molecular diversity and host associations of Posthodiplostomum in fish-eating birds across the Neotropical region of Mexico, employing an integrative taxonomic approach, we generated sequences of the large subunit (28S), internal transcribed spacers (ITS1–5.8S–ITS2) from nuclear DNA, and cytochrome c oxidase subunit 1 (cox1) from mitochondrial DNA from adult specimens of Posthodiplostomum. The main objectives of this study were to explore the molecular diversity of Posthodiplostomum in this region, to establish molecular links between newly sequenced adults and previously identified genetic lineages of metacercariae, and to expand our understanding of host and locality records for the genus.
Materials and methods
Specimen collection and morphological analyses
Seven specimens of fish-eating birds representing 2 families, Ardeidae and Laridae Rafinesque were collected in 4 localities in Mexico (Fig. 1; Table 1). Birds were identified following Howell and Webb (Reference Howell and Webb1995), and the American Ornithologist’ Union (1998). Adult diplostomids morphologically identified as Posthodiplostomum spp., were obtained from the intestines of 6 avian hosts. Diplostomids were heat-killed with distilled water, and preserved in 100% ethanol for DNA analyses. Additionally, specimens were fixed in hot 4% formalin for scanning electron microscopy studies.

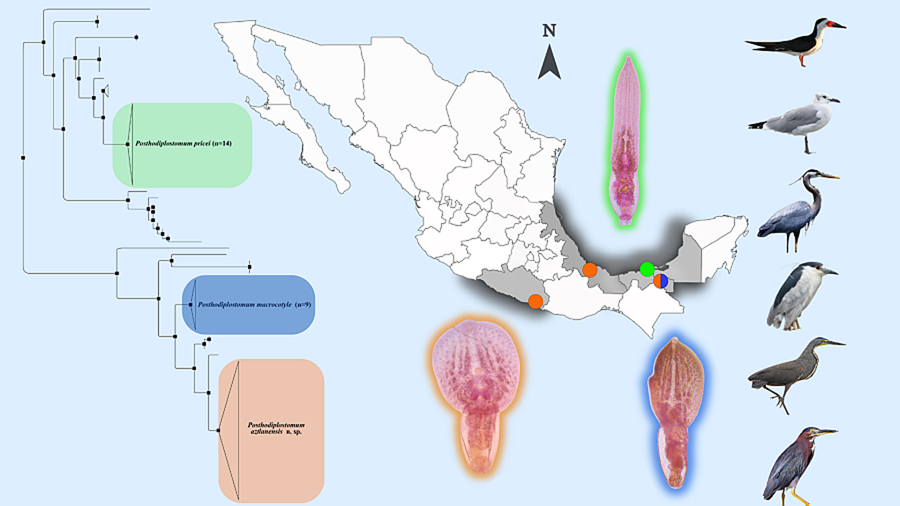
Figure 1. Sampling collection in Mexico. (1) Marquelia, Guerrero (16°35′41.5″N, 98°50′38″W), (2) Tlacotalpan, Veracruz (18°36′0″N, 95°39′0″W), (3) Nuevo Campechito, Champeche (18°38′55.849″N, 92°28′2.578″W), (4) Emiliano Zapata, Tabasco (17°46′29.1″N, 91°44′24.9″W). The colours represent the species recovered; in orange, Posthodiplostomum aztlanensis n. sp., in green Posthodiplostomum pricei and in blue Posthodiplostomum macrocotyle.
Table 1. Summary data for the taxa used in the phylogenetic analyses

Sequences in bold were obtained in this study. A (adult), M (metacercariae), C (cercaria).
a Previously included in Posthodiplostomum sp. lineage V (sensu Pérez Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022).
Specimens preserved in 100% ethanol were stained with Mayer's paracarmine (Merck, Darmstadt, Germany), dehydrated in a graded ethanol series, cleared with methyl salicylate and mounted on permanent slides with Canada balsam. Specimens were photographed and measured using a Leica DM 1000 LED compound microscope (Leica Microsystems CMS GmbH, Wetzlar, Germany); measurements are reported in micrometres (μm). Internal morphological features were illustrated using a drawing tube attached to a Leica MC120HD microscope. Drawings were made using Adobe Illustrator 27.9 (Adobe, Inc., CA, USA). Voucher specimens were deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de Mexico (UNAM), Mexico City.
Additionally, some specimens preserved in 4% formalin were dehydrated in a graded ethanol series, critical point dried, sputter-coated with gold and examined with a Hitachi Stereoscan Model S-2469N scanning electron microscope at 15 kV at LaNABIO, Instituto de Biología, UNAM.
Molecular study
Prior to extraction of the genomic DNA, specimens preserved in 100% ethanol were mounted on a microscope slide, and images were taken as references with a bright field Leica DM 1000 LED microscope (Leica, Wetzlar, Germany). Each image was linked with its genomic DNA, (photogenophore sensu Andrade-Gómez and García-Varela, Reference Andrade-Gómez and García-Varela2021). Specimens were removed from the microscope slide and genomic DNA was isolated, following the protocol described by González-García et al. (Reference González-García, Ortega-Olivares, Andrade-Gómez and García-Varela2020). The 28S, ITS1–5.8S–ITS2 and cox1 genes were amplified by polymerase chain reactions (PCR). The 28S amplifications used forward primer 391, 5′-AGCGGAGGAAAAGAAACTAA-3′ (Nadler et al., Reference Nadler, D'Amelio, Fagerholm, Berland and Paggi2000), and reverse primer 536, 5′-CAGCTATCCTGAGGGAAAC-3′ (Garcia-Varela and Nadler, Reference García-Varela and Nadler2005). The ITS amplifications used forward primer BD1 5′-GTCGTAACAAGGTTTCCGTA-3′ (Bowles and McManus, Reference Bowles and McManus1993) and the reverse primer BD2 5′-ATCTAGACCGGACTAGGCTGTG-3′ (Bowles et al., Reference Bowles, Blair and McManus1995). The cox1 gene was amplified in 2 overlapping fragments. The first region amplifications used forward primer PosthoCoiF, 5′-ATGATWTTTTTTTTYYTRATGCC-3′ and reverse primer PosthoSec1 5′-AAADGAAGAACCRAAWTTHCGATC-3′. The second region amplifications used forward JB3, 5′-TTTTTTGGGCATCCTGAGGTTTAT-3′ and the reverse primer JB4, 5′-TAAAGAACATAATGAAATTG-3′ (Bowles and McManus, Reference Bowles and McManus1993).
PCR reactions (25 μL) consisted of 1 μL of each primer (10 μ m), 2.5 μL of 10× buffer, 1.5 μL of 2 mm MgCl2, 0.5 μL of dNTPs (10 mm), 16.375 μL of water, 2 μL of genomic DNA and 0.125 μL of Taq DNA polymerase (Platinum Taq, Invitrogen Corporation, São Paulo, Brazil). PCR cycling conditions amplifications included initial denaturation at 94°C for 3 min, followed by 35 cycles of 1 min at 94°C, 1 min at 48°C for fist region of cox1, 45°C for second region of cox1 and 50°C for ITS1–5.8S rDNA–ITS2 and 28S, and 1 min at 72°C; followed by a final 10 min at 72°C. Sequencing reactions were performed using ABI Big Dye (Applied Biosystems, Boston, MA, USA) terminator sequencing chemistry and reaction products were separated and detected using an ABI 3730 capillary DNA sequencer. Contigs were assembled, base-calling differences resolved using Codoncode Aligner version 9.0.1 (Codoncode Corporation, Dedham, MA, USA) and submitted to the GenBank (Table 1).
Alignments and phylogenetic analyses
Newly generated sequences of 28S, ITS1–5.8S–ITS2 and cox1 were aligned with other diplostomid sequences available in GenBank (Table 1). Sequences of each molecular marker were aligned using SeaView version 4 (Gouy et al., Reference Gouy, Guindon and Gascuel2010) and adjusted with Mesquite program (Maddison and Maddison, Reference Maddison and Maddison2011). The nucleotide substitution model was selected using jModelTest v2.1.7 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012) applying the Akaike information criterion. The best nucleotide substitution model for 28S and ITS dataset was TVM + I + G and for both regions of cox1 was GTR + G + I.
Phylogenetic analyses were reconstructed through Bayesian inference (BI) and maximum likelihood (ML) using the online interface Cyberinfrastructure for Phylogenetic Research (CIPRES) Science Gateway v3.3 (Miller et al., Reference Miller, Pfeiffer and Schwartz2010). BI analysis was inferred with MrBayes v.3.2.7 (Ronquist et al., Reference Ronquist, Teslenko, Van Der Mark, Ayres, Darling, Höhna, Larget, Liu, Suchard and Huelsenbeck2012), with 2 simultaneous runs of the Markov Chain Monte Carlo (MCMC) for 10 million generations, sampled every 1000 generations, using a heating parameter value of 0.2 and a burn-in of 25%. ML analysis was carried out with RAxML v.7.0.4 (Silvestro and Michalak, Reference Silvestro and Michalak2011), and 1000 bootstrap replicates were run to assess nodal support. Phylogenetic trees were drawn and edited in FigTree v.1.3.1 (Rambaut, Reference Rambaut2012). Genetic divergence among taxa was estimated using uncorrected ‘p’ distances with MEGA6 (Tamura et al., Reference Tamura, Stecher, Peterson, Filipski and Kumar2013).
Results
Phylogenetic analyses
Nuclear genes
The 42 newly generated (28S) sequences were analysed together with 42 sequences of Posthodiplostomum spp. plus sequences of 6 species of diplostomids used as outgroups (Table 1). The alignment comprised 90 sequences with 1098 characters after trimming to the shortest sequence. The phylogenetic analyses identified Posthodiplostomum as a monophyletic assemblage with strong bootstrap support (100%) and a strong Bayesian posterior probability (1.0) (Fig. 2). The phylogenetic trees revealed 9 main clades (Fig. 2). The first clade contained sequences of Posthodiplostomum sp. metacercariae from the Indomalayan and Palaearctic regions. Clades II–VI formed a single lineage representing the following species: P. cuticola von Nordmann, 1832; P. brevicaudatum von Nordmann, 1832; P. nanum Dubois, Reference Dubois1937; P. minimum; and P. centrarchi Hoffman, 1958 (Fig. 2). Clade VII included sequences of P. pacificus Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021, and P. anterovarium Dronen, 1985, and 12 new sequences of adult specimens from Rynchops niger L, from Campeche, Mexico (locality 3 in Fig. 1), which nested with 2 sequences (MZ710972–MZ710973), identified as P. pricei (Krull, Reference Krull1934), from Larus delawarensis Ord., from North Dakota, USA. Clade VIII included sequences of unidentified species of Posthodiplostomum sp.; P. podicipitis Yamaguti, 1939; P. recurvirostrae Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021; P. scardinii Shulman, 1952; and P. ptychocheilus Faust, 1917. Finally, clade IX consisted of 6 subclades. One of them included 2 sequences previously identified as P. macrocotyle Dubois, Reference Dubois1937 (MZ710958–MZ710959) from Brazil nested with 7 new sequences from adult specimens (Fig. 3) (Tigrisoma mexicanum Swainson, Ardea herodias L. and Leucophaeus atricilla L.) from Tabasco, Mexico (locality 4 in Fig. 1). Another subclade included 22 newly sequenced individuals from A. herodias, Butorides virescens L, N. nycticorax L and T. mexicanum sampled in 3 localities of Mexico (localities 1, 2 and 4 in Fig. 1), plus 1 sequence from a poecilid fish from Las Brisas del Chamalecon, Honduras, identified as Posthodiplostomum sp. lineage V (sensu Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022). This clade represents a new species described herein as Posthodiplostomum aztlanensis n. sp. (Fig. 2).

Figure 2. Phylogenetic trees inferred with maximum likelihood (ML) and consensus Bayesian inference (BI) of 28S from nuclear ribosomal DNA. Numbers near internal nodes show maximum likelihood bootstrap percentage values and Bayesian posterior probabilities. Sequences generated in this study in bold. Clades highlighted in pink and blue are equivalent in the phylogenetic trees inferred with internal transcribed spacers from nuclear ribosomal DNA (Fig. 4).

Figure 3. Photogenophores of Posthodiplostomum macrocotyle. Specimens collected in Emiliano Zapata, Tabasco, Mexico from Tigrisoma mexicanum (A); Leucophaeus atricilla (B); Ardea herodias (C). Scale bars: 200 μm.
The 22 newly generated ITS sequences were analysed together with 60 sequences of Posthodiplostomum spp., plus 7 sequences from other diplostomids downloaded from the GenBank dataset that were used as outgroups (Table 1). The ITS1–5.8S–ITS2 alignment consisted of 89 sequences with 1100 characters after trimming to the shortest sequence. The phylogenetic analyses inferred with the ITS dataset also revealed the monophyly of Posthodiplostomum (Fig. 4). In particular, clade V included sequences of Posthodiplostomum sp. 8, 2 (sensu Locke et al., Reference Locke, McLaughlin and Marcogliese2010), and Posthodiplostomum sp. lineage II (sensu Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022), plus 9 new sequences of Posthodiplostomum spp. from Campeche, Mexico (locality 3 in Fig. 1), nested with 2 sequences previously identified as P. pricei (HM064959–HM064960) from the white perch (Morone americana Gmelin) from Canada (Fig. 4), showing conspecificity. Clade VIII was formed by Posthodiplostomum sp. 9 (sensu Hoogendoorn et al., Reference Hoogendoorn, Smit and Kudlai2019), Posthodiplostomum sp. lineage IV and VI (sensu Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022), and P. nanum plus 4 new sequences identified as P. macrocotyle from 3 host species (Fig. 4) (T. mexicanum, A. herodias and L. atricilla) from Tabasco, Mexico (locality 4 in Fig. 1). The sister subclade of the latter consisted of 19 sequences representing the new species from 3 localities across Mexico (including 3 sequences of metacercariae identified as Posthodiplostomum sp. lineage V (sensu Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022) from poecilids, goodeids and eleotrids from Honduras, Mexico and Costa Rica (Fig. 4).

Figure 4. Phylogenetic trees inferred with maximum likelihood (ML) and consensus Bayesian inference (BI) of ITS1–5.8S–ITS2 from nuclear ribosomal DNA. Numbers near internal nodes show maximum likelihood bootstrap percentage values and Bayesian posterior probabilities. Sequences generated in this study in bold. Clades highlighted in pink and blue colours are equivalent in the phylogenetic trees inferred with the large subunit from nuclear ribosomal DNA (Fig. 2).
Mitochondrial gene
For the cox1 gene, 2 datasets were used. The first included the cox1 barcoding region. This dataset included 5 new sequences, 80 sequences of Posthodiplostomum spp., plus 6 sequences of diplostomids as an outgroup. The alignment was 553 bp long. With ML and BI, phylogenetic analyses identified Posthodiplostomum as monophyletic, although with moderate posterior probability and low bootstrap support values. Furthermore, P. pacificus was identified as the sister taxon of an unresolved clade that included all the remaining species/lineages of Posthodiplostomum (Fig. 5A). Three sequences of the new species nested with sequences of lineage V (sensu Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022). The other 2 sequences nested with P. pricei. The second alignment included approximately 380 bp, which corresponds to the 3′ region of the cox1 gene. The topology of the tree is better resolved, although it contains a small number of sequenced individuals. This dataset contained 4 new sequences, 12 sequences of Posthodiplostomum spp. plus 8 diplostomids used as an outgroup. The tree also revealed the monophyly of Posthodiplostomum as well as the monophyly of the 4 new sequences; 2 belonged to P. pricei, and the other 2 corresponded to the new species (Fig. 5B).

Figure 5. Phylogenetic trees inferred with maximum likelihood (ML) and consensus Bayesian inference (BI) mitochondrial cytochrome c oxidase subunit 1 (cox1) genes. The first region of the cox1 (A). The second region of the cox1 (B). Numbers near internal nodes show maximum likelihood bootstrap percentage values and Bayesian posterior probabilities.
Genetic divergence
The 28S intraspecific genetic divergence among 14 isolates of P. pricei was very low, ranging from 0 to 0.09%, whereas that among 9 isolates of P. macrocotyle ranged from 0 to 0.18%, and that among 23 isolates of P. aztlanensis n. sp. ranged from 0 to 0.45% (Supplementary Table 1). The interspecific divergence among Posthodiplostomum spp. varied between 0 and 7.86%; the greatest divergence was found between 1 isolate of P. macrocotyle from L. atricilla in Tabasco, Mexico, and Posthodiplostomum sp. 1 from Trichopodus trichopterus Pallas, in Vietnam (MT394051). The interspecific divergence between the new species and all congeners varied from 0.63 to 5.23%.
The intraspecific genetic divergence of the ITS region among the 11 P. pricei isolates was also low, ranging from 0 to 0.78%; the greatest difference was found between 1 isolate (HM064959) from M. americana in Canada and 1 isolate from R. niger in Campeche (locality 3 in Fig. 1), whereas the divergence among the 4 P. macrocotyle isolates ranged from 0 to 0.11% and among the 21 P. aztlanensis n. sp. isolates ranged from 0 to 0.38%. The interspecific genetic divergence of the ITS region between the new species and all other species ranged from 1.18 to 11.7% (Supplementary Table 2).
Finally, the cox1 intraspecific genetic divergence among isolates of P. pricei ranged from 0 to 2.6%, and among isolates of P. aztlanensis n. sp., the divergence varied from 0.47 to 0.94% and 0.53% from the first and second regions of cox1, respectively. For the interspecific genetic divergence of cox1, 2 values were obtained, 1 for each database (Supplementary Tables 3 and 4). The largest interspecific genetic divergence for the first region of cox1 of Posthodiplostomum spp. ranged from 19 to 22.3% between P. pricei and P. cuticola, whereas for the second region of cox1, it ranged from 18.6 to 19.3% between P. aztlanensis and Posthodiplostomum lineage II.
Morphological description
Family Diplostomidae Poirier, 1886
Genus Posthodiplostomum Dubois, 1936
Posthodiplostomum aztlanensis n. sp.
Type host: Butorides virescens (Little Green Heron) (Pelecaniformes: Ardeidae).
Other hosts: Ardea herodias (Great Blue Heron) (Ardeidae); Nycticorax nycticorax (black-crowned Night Heron) (Ardeidae); T. mexicanum (bare-throated Tiger-Heron) (Ardeidae).
Type locality: Marquelia, Guerrero, Mexico (16°35′41.5″N, 98°50′38″W).
Other localities: Emiliano Zapata, Tabasco, Mexico (17°46′29.1″N, 91°44′24.9″W); Tlacotalpan, Veracruz, Mexico (18°36′0″N, 95°39′0″W).
Site in host: Intestine
Type material: Holotype CNHE: 12990; paratypes CNHE: 12991–12993
GenBank accession number: 28S: PP718597–PP718619; ITS: PP718639–718656; cox1: PP724755–PP724757.
ZooBank registration: To comply with the regulations set out in article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012), details of the new species have been submitted to ZooBank. The Life Science Identifier (LSID) for P. aztlanensis n. sp. is urn:lsid:zoobank.org:act:E12C7AD7-BB6D-411B-A3BC-43D38ADB71C0
Etymology: The epithet is dedicated to the city of ‘Aztlan’, where the Aztec culture originated, which in Nahuatl means place of herons.
Description (Fig. 6; Table 2)
Description (based on 33 adult specimens); measurements of holotype (Fig. 6B) given in text; measurements of the entire series given in Table 2. Body 1096 long, consisting of distinct prosoma and opisthosoma (Fig. 6C); prosoma oval, 676 long, widest at mid-length, 614 wide. Opisthosoma cylindrical, 502 long, much narrower than prosoma, 276 wide. Prosoma: opisthosoma length ratio 1:1.3. Tegument completely armed with pectinate spines (Fig. 6D). Oral sucker terminal, 48 (length) × 54 (width). Ventral sucker equal size to oral sucker, 46 × 54, post equatorial of prosoma. Oral: ventral sucker ratio 1:1.05 × 1:0.98. Holdfast organ immediately posterior to ventral sucker, oval transversely elongated with ventral muscular portion, 157 × 201. Proteolytic gland dorsal to posterior part of holdfast organ, bilobed. Prepharynx not observed. Pharynx oval, 46 × 36. Oesophagus larger than pharynx, 55 long. Caecal bifurcation in the most anterior quarter of prosoma length. Caeca slender, end not observed due to vitellarium.

Figure 6. Posthodiplostomum aztlanensis n. sp. collected from Butorides virescens in Marquelia, Guerrero, Mexico. Ventral view of the photogenophore (A); photograph of the holotype (B); ventral view of the holotype (C); scanning electron micrograph, tegument spines (D); posterior end of the holotype and genital cone (E); whole worm (F). Scale bars: (A, B, C) 200 μm; (D) 10 μm; (E) 20 μm; (F) 400 μm.
Table 2. Comparative measurements of adult specimens of Posthodiplostomum aztlanensis n. sp. and Posthodiplostomum pricei

Measurements in micrometres. aHost experimental. bEstimated from the published drawing (Krull, Reference Krull1934).
Testes 2, tandem; anterior testis positioned posterior to prosoma end, subspherical 149 × 141, posterior testis somewhat bilobed, 163 × 167. Seminal vesicle post-testicular, ventral to posterior testis, compact, continues to short ejaculatory duct. Ejaculatory duct joins metraterm to form hermaphroditic duct. Hermaphroditic duct opening at genital cone into genital atrium; genital cone surrounded by prepuce within genital atrium. Genital pore terminal (Fig. 6E).
Ovary pretesticular, posterior part of ovary ventral to anterior testis, transversely oval, positioned near prosoma–opisthosoma junction and posterior to proteolytic gland, 70 × 92. Oötype and Mehlis' gland not observed. Laurer's canal not observed. Vitellarium located from near caecal bifurcation in prosoma, extending to opisthosoma to the posterior margin of testis. Eggs not observed. Excretory vesicle not observed. Excretory pore subterminal.
Remarks
Posthodiplostomum aztlanensis n. sp. belongs to genus Posthodiplostomum based on the results of our molecular analyses as well as the presence of a genital prepuce and lack of pseudosuckers. The new species can be distinguished from all other Posthodiplostomum spp., except for Posthodiplostomum biellipticum Dubois, 1958 and Posthodiplostomum grayi (Verma, 1936), by its prosoma shape (oval), whereas variable form in all other Posthodiplostomum spp. (concave, linguiform or lanceolate). The new species and P. biellipticum can be further distinguished based on the prosoma: opisthosoma length ratio (opisthosoma being longer in P. biellipticum than P. aztlanensis). In addition, both species P. biellipticum and P. grayi can be distinguished based on length of body (1450 in P. biellipticum and P. grayi, vs 779–1392 in P. aztlanensis). The biogeographical distribution can be used as another character to distinguish the species. For example, P. biellipticum has been recorded in Ghana (Africa), P. grayi in India, China, Philippines (Asia), whereas P. aztlanensis was recorded in the Neotropical region of Mexico (Americas).
Morphological identification
Posthodiplostomum pricei (Krull, Reference Krull1934)
Host: Rynchops niger (Black Skimmer) (Charadriiformes: Laridae).
Locality: Nuevo Campechito, Campeche, Mexico (18°38′55.849″N, 92°28′2.578″W).
Site in host: Intestine
Voucher: CNHE: 12994
GenBank accession number: 28S:PP718620–PP718631; ITS: PP718657–PP718665; cox1: PP724758–PP724759.
Sixteen adult specimens were collected, measured and compared with described species. Our specimens were morphologically identified as P. pricei; overall, specimens are similar to those described of P. pricei by Krull (Reference Krull1934) in the original description, and redescribed later by Dubois (Reference Dubois1970). In addition, the genetic data generated in this study supported the morphological evidence, confirming that all the specimens belong to P. price. Our specimens are similar to those descriptions for the prosoma shape (lanceolate), ovary position (intertesticular), the prosoma:opisthosoma length ratio, prosoma:body length ratio, the holdfast:prosoma length ratio, the oral sucker:pharynx length ratio and the oesophagus length. Our specimens are, however, smaller than those from previous descriptions (Fig. 7; Table 2).

Figure 7. Posthodiplostomum pricei collected from Rynchops niger in Nuevo campechito, Campeche, Mexico, ventral view of the photogenophore (A); photograph of the vouchers (B); ventral view (C); scanning electron micrograph, oral sucker and tegument spines (D); posterior end of the voucher and genital cone (E); whole worm (F). Scale bars: (A, B, C, F) 200 μm; (D) 10 μm; (E) 20 μm.
Discussion
Adults of the genus Posthodiplostomum are known to infect the intestines of fish-eating birds, mainly those of the family Ardeidae (Ritossa et al., Reference Ritossa, Flores and Viozzi2013; López-Hernández et al., Reference López-Hernández, Locke, De Melo, Rabelo and Pinto2018; Perez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022; Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021). López-Hernández et al. (Reference López-Hernández, Locke, De Melo, Rabelo and Pinto2018) suggested that species of Posthodiplostomum have diversified in the Neotropical region. More recently, Pérez-Ponce de León et al. (Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022) assessed the diversity of the genus through an analysis of the genetic variation of metacercariae in freshwater fishes across Middle America (Mexico, Guatemala, El Salvador, Honduras and Costa Rica). These authors sequenced 2 molecular markers, the internal transcribed spacer (ITS1–5.8S–ITS2) and 1 region of the mitochondrial cox1 gene. Their molecular analyses yielded 6 genetic lineages that did not correspond to any available sequences of Posthodiplostomum in GenBank at the time. Finally, Pernett et al. (Reference Pernett, Brant and Locke2022) suggested that the biodiversity of Posthodiplostomum in the Neotropical region was sub estimated. In the present study, adult specimens were collected from fish-eating birds at several locations in the Neotropical region of Mexico. Phylogenetic analyses with 28S, ITS and cox1 revealed that adults were allocated to 3 independent clades. One of these clades corresponded to lineage V (sensu Pérez-Ponce de León et al., Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022), and we linked the metacercariae recovered from 3 fish families (Goodeidae Jordan, Eleotridae Bonaparte and Poecilidae Bonaparte). This lineage represented a new species, P. aztlanensis n. sp., which seems to be, as adults, host-specific to birds of the family Ardeidae. This represents the first species described in the Neotropical region of Mexico. In addition to morphological evidence and the position of the new lineage in the phylogenetic trees, the genetic divergence found between adults and metacercariae provided additional support for the separation of the species. For example, the intraspecific genetic divergence among isolates was very low (0–0.45% for 28S, 0–0.38% for ITS, 0.47–0.94% and 0.53% for the first and second regions of cox1). This low divergence level, particularly that of cox1, is similar to that reported previously by Achatz et al. (Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021) (less than 4.1% were considered conspecific). The interspecific divergence between the new species and its congeners varied from 0.63 to 5.23% for 28S, 1.18 to 11.7% for ITS, and 10.3 to 19.4% and 10.9 to 19.3% for the first and second regions of cox1, respectively. These range values are larger than those previously reported by Achatz et al. (Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021), which were 4.1%.
Furthermore, molecular analyses were useful for identifying 2 additional species of Posthodiplostomum. One of them was P. macrocotyle, which was found in 3 bird species (T. mexicanum, A. herodias and L. atricilla) from Tabasco, Mexico (see Fig. 3); these records represent new locality records and expand the distribution range of the species. Newly generated sequences were placed together in a clade with 2 sequences identified as P. macrocotyle from the black-collared hawk (Busarellus nigricollis Latham) from Brazil (MZ710958–MZ71095, Fig. 2), with a low genetic divergence value (0–0.18%). Posthodiplostomum macrocotyle was originally described by Dubois (Reference Dubois1937) from specimens recovered from the black skimmer R. niger in Brazil. Therefore, the presence of P. macrocotyle expands the geographical distribution of the species further north in the Neotropical region. Moreover, P. macrocotyle is considered a generalist species since it has been recorded in at least 5 host species belonging to 3 bird families (Accipitridae Vieillot, Laridae and Ardeidae). However, no matches were found between P. macrocotyle and the genetic lineages of metacercariae reported in Pérez-Ponce de León et al. (Reference Pérez-Ponce de León, Sereno-Uribe, Pinacho-Pinacho and García-Varela2022).
The second species, supported by phylogenetic analyses, genetic divergence and morphological evidence, corresponded to P. pricei. The taxonomic history of this taxon has been controversial. The species was originally described as Neodiplostomum pricei by Krull (Reference Krull1934) as a parasite of the silver gull Chroicocephalus novaehollandiae Stephens in Washington, USA; the species was later transferred to the genus Mesoophorodiplostomum by Dubois (Reference Dubois1936) and accepted by Niewiadomska (Reference Niewiadomska, Gibson, Jones and Bray2002). The first sequences of metacercariae from 3 fish species (Fundulus diaphanous Lesueur, F. heteroclitus L. and Lepomis gibossus L.) from Canada were assigned to Posthodiplostomum sp. 6 (Moszczynska et al., Reference Moszczynska, Locke, McLaughlin, Marcogliese and Crease2009; Locke et al., Reference Locke, McLaughlin and Marcogliese2010). Later, a sequence from an adult specimen experimentally obtained from the American herring gull (Larus argentatus Pontoppidan) was identified as P. pricei (see Blasco-Costa and Locke, Reference Blasco-Costa and Locke2017). More recently, Achatz et al. (Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021) obtained sequences (28S and cox1) from an adult specimen recovered from the ring-billed gull L. delawarensis in North Dakota, USA. Their phylogenetic analyses placed M. pricei within the genus Posthodiplostomum and transferred M. pricei to Posthodiplostomum as P. pricei (Krull, Reference Krull1934). Additionally, the sequences of metacercariae, referred to as Posthodiplostomum sp. 6, were linked with those sequences of Blasco-Costa and Locke (Reference Blasco-Costa and Locke2017) and transferred to P. pricei (see Achatz et al., Reference Achatz, Chermak, Martens, Pulis, Fecchio, Bell, Greiman, Cromwell, Brant, Kent and Tkach2021). Our specimens from the black skimmer, R. niger L., which were sampled in Campeche, Mexico, match all these sequences and expand southwards the distribution range of the species from the Nearctic region to southeastern Mexico in the Neotropical region. In this case, P. pricei shows narrow host specificity towards its definitive host (Laridae).
Therefore, considering the 3 species reported in this study, in addition to at least 5 other genetic lineages (candidate species) of the genus Posthodiplostomum occurring in Mexico, we could consider it a hotspot of diversity due to its transitional position between the Nearctic and Neotropical biogeographical regions (Morrone, Reference Morrone2006; Pérez-Ponce de León et al., Reference Pérez-Ponce de León, García-Prieto and Mendoza-Garfías2007). In addition, the results of the present study suggest that the Neotropical region of Mexico meets the ecological requirements to complete the life cycle of P. aztlanensis n. sp., P. macrocotyle and P. pricei, which is key to their distribution. The same pattern of sympatric distribution has been observed in other species of diplostomids, strigeids and clinostomids. For example, Tylodelphys aztecae (García-Varela et al., Reference García-Varela, Sereno-Uribe, Pinacho-Pinacho, Hernández-Cruz and Pérez-Ponce de León2016) was found in the Neotropical region of Mexico, whereas Tylodelphys sp. 6 (sensu Locke et al., Reference Locke, Al-Nasiri, Caffara, Drago, Kalbe, Lapierre, McLaughlin, Nie, Overstreet, Souza, Takemoto and Marcogliese2015) was initially recorded in the Nearctic region and was later found in the Neotropical region of Mexico (Sereno-Uribe et al., Reference Sereno-Uribe, Andrade-Gómez, De León and García-Varela2018); Strigea macrobursa (Drago and Lunaschi, 2011) was described in Argentina, and it has been recorded in Mexico, together with Strigea magnirostris (López-Jiménez et al., Reference López-Jiménez, González-García, Andrade-Gómez and García-Varela2023). Similarly, Clinostomum tataxumui is restricted to the Neotropical region, whereas Clinostomum marginatum has been recorded in both the Nearctic and Neotropical regions (Sereno-Uribe et al., Reference Sereno-Uribe, Pinacho-Pinacho, García-Varela and Pérez-Ponce De León2013).
Finally, our study represents a step forward in our comprehension of parasite biodiversity in biogeographical transitional areas and provides new molecular and morphological data to delineate and describe new species of trematodes infecting fish-eating birds. Nevertheless, a larger bird sampling effort is required to increase the genetic library of the trematodes infecting birds to establish a more precise link with the metacercariae found in a diverse array of fish.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024000970
Data availability statement
The genetic distances estimated among the taxa for each molecular marker can be download. The alignments can be obtained from the corresponding author upon request.
Acknowledgements
This paper serves as fulfilment of M. T. G.-G. for obtaining an M.Sc. degree in the Posgrado en Ciencias Biológicas, UNAM. We thank the Consejo Nacional de Humanidades, Ciencias y Tecnologías CONAHCYT for funding and for the support of this research through a graduate scholarship to M. T. G.-G. (CVU 956064). We also thank Berenit Mendoza for her help with the use of the SEM unit and Laura Márquez and Nelly López Ortiz from LaNabio for their help during the sequencing of the DNA fragments.
Author contributions
M. T. G.-G., G. P.-P. d. L. and M. G.-V. conceived and designed the study. M. T. G.-G., A. L.-J., A. L. S.-U. and M. P. O.-O. conducted data gathering. M. T. G.-G. and A. L.-J. performed phylogenetic analyses. M. T. G.-G., G. P.-P. d. L. and M. G.-V. wrote and edited the article.
Financial support
This research was supported by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT-UNAM) IN201122 to M. G.-V. and IN200824 to G. P.-P. d. L.
Competing interests
None.
Ethical standards
The sampling in this work complies with the current laws and animal ethics regulations of Mexico. Specimens were collected under the Cartilla Nacional de Colector Científico (FAUT 0202) issued by the Secretaría del Medio Ambiente y Recursos Naturales (SEMARNAT), to M. G.-V.