INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) infections in persons lacking established healthcare-associated (HA)-MRSA risk factors were first reported in the 1990s. Community-associated (CA)-MRSA infections have now been observed throughout the USA [Reference Frank1–Reference Adcock3] and worldwide [Reference Salmenlinna, Lyytikainen and Vuopio-Varkila4, Reference Turnidge and Bell5] and the burden of disease has increased rapidly [Reference Kaplan6, Reference Moran7]. Outbreaks and clusters of CA-MRSA infections have been reported in a variety of settings [Reference Adcock3, 9–Reference Saiman12]. Risk factors for CA-MRSA infection or colonization identified in outbreaks include close person-to-person contact, non-intact skin, suboptimal hygiene, and prior antimicrobial use [9, Reference Baggett13–Reference Turabelidze16].
Despite the increasing incidence of non-outbreak CA-MRSA, little is known about risk factors for CA-MRSA infections in the general population. To determine risk factors for CA-S. aureus (CA-SA) and CA-MRSA in the general population, the Minnesota Department of Health (MDH) initiated a hypothesis-generating case-control study of CA-SA infections. In this study we conducted a case-case comparison of CA-MRSA to CA-methicillin sensitive S. aureus (MSSA) cases as well as a case-control comparison of CA-MRSA cases to matched uninfected community controls and CA-MSSA cases to matched uninfected community controls.
METHODS
The MDH established a prospective MRSA surveillance network of 12 hospital sentinel laboratories in 2000 that has been described elsewhere [Reference Naimi17]. Cases of laboratory-confirmed MRSA were reportable to MDH and isolates were submitted to the MDH Public Health Laboratory (PHL). Based on the number of CA-MRSA cases reported in 2000–2002, seven sentinel sites that reported at least 10 CA-MRSA cases per year were targeted for participation in the study. Four sites agreed to participate in the study; two paediatric facilities located in the Minneapolis-St Paul Metropolitan Area and two facilities in greater Minnesota that serve paediatric and adult patients. All participating sites serve in-patients, outpatients, and are reference laboratories for other clinical practices.
CA-MRSA case-patients were defined as patients with clinically relevant MRSA infections who lacked the following established HA-MRSA risk factors according to the Centers for Disease Control and Prevention (CDC) case definition: MRSA identified after 48 h of admission; history of hospitalization (excluding infant hospitalization for an uncomplicated birth), surgery, dialysis, or residence in a long-term care facility within 1 year prior to the MRSA culture date; a permanent indwelling catheter or percutaneous device at the time of culture; or known previous MRSA infection or colonization. To be eligible for the study, CA-MSSA cases were required to meet the same criteria as CA-MRSA cases regarding lack of HA-MRSA risk factors, with the exception of prior S. aureus infection.
CA-MRSA cases were identified by hospital infection preventionists (IPs) who reviewed medical records to determine if patients met the CA-MRSA case definition. Cases were grouped into eight age groups: 0 to <6 months, 6–23 months, 2–4 years, 5–11 years, 12–17 years, 18–40 years, 40–64 years, and ⩾65 years. Culture sites were grouped into three categories: invasive infections (e.g. blood, joint, bone), skin or soft tissue infections (SSTIs), and non-SSTI/non-invasive (e.g. ear, eye, urine). For each CA-MRSA case, IPs attempted to identify five age-group and culture site-matched CA-MSSA cases. If a matched CA-MSSA case could not be identified during the study period the CA-MRSA case was excluded from the analysis. For each CA-MRSA and CA-MSSA case we attempted to obtain three age groups and geographically matched uninfected community controls; sets with two controls enrolled were included in the analysis. For cases aged <2 years, birth records were used to identify potential controls using the same birth date and zip code as the case. For patients aged ⩾2 years, controls were ascertained by sequential digit dialling of telephone numbers. Community controls were required to meet criteria regarding lack of HA-MRSA risk factors with the exception of prior S. aureus infections. If more than 250 residential numbers were called and at least two controls could not be enrolled, the case was not included in the analysis. The study continued until 75 sets of participants were enrolled (i.e. one CA-MRSA case and at least two CA-MRSA uninfected community controls; one CA-MSSA case and two CA-MSSA uninfected community controls), amounting to 6–8 participants per set; only complete sets were included in the analysis. For cases, enrolment began 1 February 2003 and was completed 15 June 2005; community control enrolment began on 1 April 2003 and was completed 30 January 2006.
Cases and controls were interviewed by study investigators to confirm study eligibility and obtain information regarding their health history and potential risk factors. For uninfected community controls, the matched case's culture date was used as the reference date. To determine possible risk factors for CA-MRSA and CA-MSSA infections, questions were asked regarding demographic and household characteristics as well as recreational, social, and bathing practices. Participants were also asked questions about their health history and the health history of household members. For participants who were aged <18 years, highest education level of the parent or guardian who was interviewed was used as a proxy for educational level. Participants were asked to identify their race and ethnicity according to categories used by the 2000 U.S. Census Bureau.
To obtain antimicrobial histories, participants were asked to identify their primary healthcare providers and all other healthcare providers visited in the 6 months prior to the reference date. Healthcare providers supplied information on antimicrobial use, including antimicrobial name and date of prescription for all antimicrobials prescribed in the relevant time period. Cases and controls for whom an antimicrobial history could not be obtained were not included in the analysis of antimicrobial use but were included in the analysis of other risk factors. Antimicrobials prescribed in the 0–30 days prior to culture were not included because we did not ascertain the reason for prescribing and could not exclude the possibility that the reason for the antimicrobial was treatment of the index S. aureus infection prior to obtaining cultures.
Consent was required for participation in the study for CA-MSSA cases and MSSA isolates, controls and a separate consent was required to obtain antimicrobial history. The study was approved by Institutional Review Boards at the MDH, CDC and participating hospitals.
Laboratory testing
MDH-PHL confirmed and characterized study isolates received. Identification of S. aureus was confirmed with a tube coagulase test (Difco Laboratories, USA), oxacillin resistance was confirmed using an oxacillin agar screen test (Becton Dickinson, USA) and antimicrobial susceptibility testing was performed by broth microdilution panel (PML Microbiologicals, USA). All testing was performed in accordance with Clinical and Laboratory Standards Institute standards [18, 19].
Molecular typing of isolates was performed by pulsed-field gel electrophoresis (PFGE) of genomic DNA after restriction endonuclease digestion with SmaI. PFGE patterns were compared using Bionumerics Software (Applied Maths, Belgium) and the Dice coefficient with optimization of 0·50% and position tolerance of 1·00%. Patterns that had exact matches of all bands in the range of 70–700 kb were considered indistinguishable. CA-MRSA isolates at least 80% similar to the pulsed-field type (PFT) reference strain were considered to be part of that PFT [Reference Tenover20, Reference McDougal21].
Polymerase chain reaction (PCR) was used to determine the presence of Panton–Valentine leukocidin (PVL), toxic shock syndrome toxin – 1 (TSST-1), and staphylococcal enterotoxin genes SEA, SEB, SEC, SED, SEK, and SEQ. One loopful of culture from TSA blood agar plate or glycerol stock was added to 200 μl DNAzol (Molecular Research Corporation, USA) solution and boiled for 15 min. The suspernatant was transferred to a Qiagen PCR Purification column, and the manufacturer's instructions were followed. An iCycler (Bio-Rad, USA) thermocycler was used for amplification with the following conditions: 95°C for 15 min, 35 cycles of 95°C for 1 min, 52°C for 1 min and 72°C for 1 min, with a final extension at 72°C for 10 min. PCR primers for PVL [Reference Lina22], SEA-D [Reference Monday and Bohach23], SEK [Reference Chiang24], SEQ [Reference Chiang25] and TSST-1 [Reference Monday and Bohach23] were as previously described. A TSST-1 primer sequence was designed to include more recently published sequences. The TSST-1 primer sequence was 5′-ATT CCT TAG GAT CTA TGC GTA T-3′. The PCR products were analysed by electrophoresis with 2% agarose gels and viewed with ethidium bromide stain.
Statistical methods
Three separate analyses were conducted: (1) comparison of CA-MRSA cases to matched CA-MSSA cases; (2) comparison of CA-MRSA cases to matched CA-MRSA community controls; and (3) comparison of CA-MSSA cases to matched CA-MSSA community controls. Univariate analysis evaluated the difference between CA-MRSA and CA-MSSA isolates using Epi Info version 6.04d (CDC, USA) and were not analysed as matched sets.
Multivariate stepwise conditional logistic regression of potential risk factors was conducted using SAS version 9.1 (SAS Institute Inc., USA) proc glm. First, a stepwise model was analysed to determine which confounding socio-economic variables to control for in the model with possible risk factors. Socio-economic variables included were income, education, and race (white vs. non-white). After evaluating fit, income and education variables were grouped and included in the multivariate model as continuous variables. Socio-economic variables with an α of ⩽0·10 were entered into the model to analyse risk factors. Possible risk factors with an α of ⩽0·10 in the univariate analysis and interaction terms were further tested in the second multivariate model, again using stepwise conditional logistic regression.
RESULTS
During the study period, a total of 246 CA-MRSA cases were identified as eligible through medical record review. One hundred and sixty CA-MRSA cases were enrolled in the study and completed interviews; 42 (17%) CA-MRSA cases could not be reached, 22 (9%) cases refused study participation, and an additional 22 (9%) cases were reclassified as HA-MRSA after answering study eligibility questions. Interviews were completed with 90 CA-MSSA cases. Because consent was obtained by participating hospitals prior to MDH contacting CA-MRSA cases, the total number of potentially eligible CA-MSSA cases who refused participation or were lost to follow-up was unknown. Eighty-five CA-MRSA cases and 15 CA-MSSA cases were enrolled in the study but were not included in the analysis because 75 sets were already identified and controls were not obtained for these cases.
A total of 75 sets that included 75 CA-MRSA cases, 75 CA-MSSA cases, and 438 uninfected community controls (226 CA-MRSA community controls and 212 CA-MSSA community controls), were analysed. Eighty percent of cases had SSTIs, 13% had non-SSTI/non-invasive infections and the remaining 7% of cases had invasive infections. The average age of CA-MRSA cases was 24 years (range 8 days to 87 years); 48% of cases were identified by paediatric hospitals; age group-matched CA-MSSA cases and controls had an average age of 22 years (range 7 days to 95 years).
Results of demographic, recreational and household characteristics for study participants are reported in Table 1. Antimicrobial use history in the 6 months prior to culture date (or reference date for community controls) were obtained from healthcare providers for 97% of CA-MRSA cases, 93% of CA-MSSA cases, 75% of uninfected CA-MRSA community controls, and 73% of uninfected CA-MSSA community controls. Responses to medical history questions are displayed in Table 2.
Table 1. Demographic, recreational, and household characteristics of CA-MRSA and CA-MSSA cases, and uninfected controls
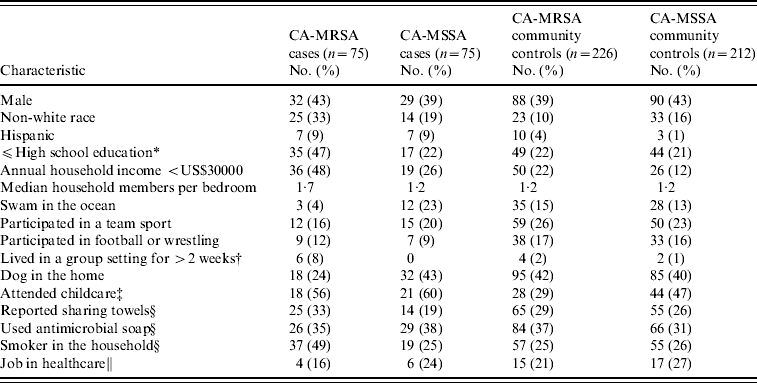
* If participant is aged <18 years, highest level of education of parent/guardian.
† Group settings included shelters, military barracks, dormitories, and prisons.
‡ Number (%) for participants aged ⩽9 years only.
§ Reference period of 1 year prior to culture date for cases and 1 year prior to culture date of matched case for community controls.
∥ Reference period 5 years prior to culture date for case and 5 years prior to culture date of matched case for community controls; number (%) for participants aged ⩾16 years only.
Table 2. Clinical characteristics of CA-MRSA and CA-MSSA cases, and uninfected controls
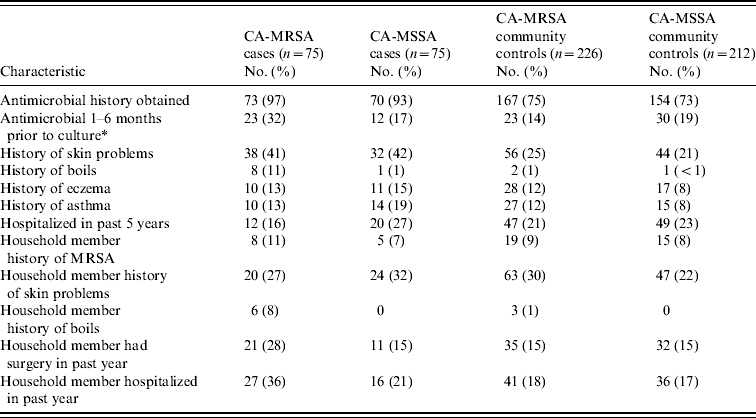
* For community controls, culture date of the matched case was used as the reference date.
Non-white race and other socio-economic variables were found to be associated with staphylococcal infections. CA-MRSA cases were more likely to be non-white race, have lower educational levels, lower household incomes, and an increased number of household members per bedroom than CA-MSSA cases or uninfected CA-MRSA community controls. CA-MRSA infections were also found to be associated with certain household practices and characteristics compared to CA-MRSA cases and community controls. CA-MSSA cases were more likely to be non-white, have Hispanic ethnicity, have lower incomes, attend childcare or have lived in group settings than were uninfected CA-MSSA community controls. CA-MRSA and CA-MSSA cases more frequently reported a history of skin problems and boils compared to community controls; CA-MRSA infections were also associated with having an antimicrobial prescribed in the 1–6 months prior to culture compared to both CA-MSSA cases and community controls (Table 3).
Table 3. Univariate analysis results for CA-MRSA and CA-MSSA cases, and uninfected controls
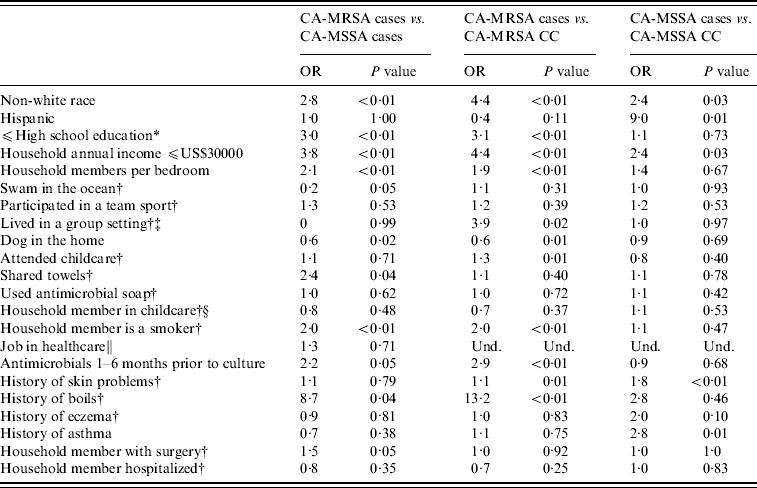
OR, Odds ratio; CC, uninfected community control; Und, undefined.
* If participant is <18 years old, highest level of education of parent/guardian.
† In the year prior to culture or reference date.
‡ Group settings included shelters, military barracks, dormitories, and prisons.
§ Number (%) for participants aged ⩽9 years only.
∥ Reference period 5 years prior to culture date for case and 5 years prior to culture date of matched case for community controls; number (%) for participants aged ⩾16 years only.
Multivariate analyses of CA-MRSA cases compared to CA-MSSA cases and community controls were adjusted for income and education (Table 4). In both models, history of antimicrobial use in the 1–6 months prior to the reference date was associated with CA-MRSA cases compared to CA-MSSA cases and uninfected CA-MRSA community controls. Additionally, CA-MRSA cases were more likely to have a history of boils and have a smoker in the home than were uninfected CA-MRSA community controls. When adjusted for race, a history of skin problems was associated with an increased risk of CA-MSSA infection compared to uninfected CA-MSSA community controls.
Table 4. Multivariate analysis results for CA-MRSA and CA-MSSA cases, and uninfected controls
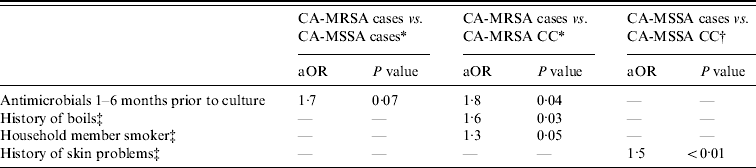
aOR, Adjusted odds ratio; CC, uninfected community control; n.s., not significant defined as P>0·10.
* Controlled for income and education.
† Controlled for race.
‡ In the year prior to culture or reference date.
MDH received isolates for 69 (92%) of CA-MRSA cases and 53 (71%) of CA-MSSA cases. Culture site of isolates received was similar to the distribution of culture sites in all cases. Of the CA-MRSA isolates received, 77% were from SSTIs, 17% from non-SSTI/non-invasive sites and 6% from invasive sites; of the CA-MSSA isolates received 81% were from SSTIs, 15% from non-SSTI/non-invasive sites and 4% from invasive sites. Molecular differences were observed between CA-MRSA and CA-MSSA isolates and the distribution of PFTs and toxin genes for CA-MRSA and CA-MSSA isolates are shown in Tables 5 and 6, respectively. Forty-nine (71%) CA-MRSA isolates carried genes for PVL and SEQ compared to two (3%) CA-MSSA isolates (OR 41·2, P<0·01). Thirty-one (45%) CA-MRSA isolates carried genes for PVL, SEQ, and SEK whereas only one (2%) CA-MSSA isolate carried these toxin genes (OR 43·2, P<0·01). TSST-1 and SEB toxins were less likely to be present in CA-MRSA isolates compared to CA-MSSA isolates (OR 0·04, P<0·01 and OR 0·20, P=0·04, respectively). Only three (4%) CA-MRSA isolates did not contain any of the toxin genes tested; 14 (26%) CA-MSSA isolates did not contain any of the toxin genes tested (OR 0·03, P<0·01).
Table 5. USA genotypes of CA-MRSA and CA-MSSA isolates
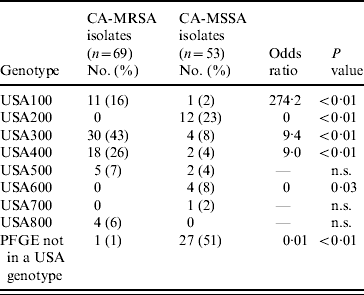
n.s., Not significant.
Table 6. Toxin profiles of CA-MRSA and CA-MSSA isolates
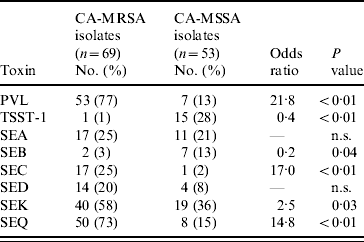
PVL, Panton–Valentine leukocidin; TSST-1, toxic shock syndrome toxin-1; SE, staphylococcal enterotoxin; n.s., not significant.
Seventy percent (48/69) of CA-MRSA isolates were USA300 (n=30) or USA400 (n=18), PFTs that are typically associated with CA-MRSA, compared to 11% (6/54) of CA-MSSA isolates. For CA-MRSA and CA-MSSA isolates combined, USA300 and USA400 isolates were more likely to contain the PVL gene (OR 74·6, P<0·01 and OR 7·8, P<0·01, respectively) compared to other isolates. USA400 isolates were more likely than other isolates to have genes for SEA and SEC toxins (OR 30·0, P<0·01 and OR 572·3, P<0·01); whereas USA300 isolates were less likely to contain genes for SEA or SEC toxins (OR 0, P<0·01 for both toxins) and less likely to contain the TSST-1 toxin gene (OR 0, P=0·02). USA100 and USA200 isolates, PFTs that are typically associated with HA-MRSA strains, were less likely than isolates of other PFTs to contain PVL genes (OR 0·01, P<0·01 for both genotypes). USA100 isolates were more likely to contain SED toxin genes (OR 8·2, P<0·01) than isolates from other PFTs and USA200 isolates were more likely to have SEA (OR 3·9, P=0·05) and TSST-1 toxin genes (OR 51·0, P<0·01) compared to isolates from other PFTs.
DISCUSSION
To the best of our knowledge, this is the first risk-factor study of S. aureus infection in non-outbreak cases that compared CA-MRSA and CA-MSSA infections and used two control groups to analyse potential risk factors associated with CA-MRSA and CA-MSSA infections. We found that CA-MRSA infections were associated with prior antimicrobial use compared to both CA-MSSA cases and uninfected community controls. To provide further support for the association between prior antimicrobial use and CA-MRSA infection, prior antimicrobial use was not associated with CA-MSSA when compared to uninfected community controls. This was in contrast to a history of skin problems, which was a risk factor for both CA-MRSA and CA-MSSA infections. It is possible that prior antimicrobial use might provide positive selective pressure for MRSA strains.
Prior antimicrobial use as a risk factor for CA-MRSA has been examined by other studies. However, because of differences in the populations studied, control groups, and methods it is difficult to compare findings. Studies from outbreak settings have found that prior antimicrobial use was associated with CA-MRSA infection and CA-MRSA colonization [Reference Baggett13–Reference Kazakova15]. However, these studies examined risk factors for CA-MRSA outbreak-related infections in defined populations. A few studies have been performed comparing prior antimicrobial use in CA-MRSA cases to CA-MSSA cases. A study in Los Angeles County found no association between prior antimicrobial use and CA-MRSA but did find that recent incarceration (Los Angeles County was experiencing a large county jail outbreak at the time) and use of illicit drugs increased risk [Reference Miller26]. Although our study and the Los Angeles County study are similar in ascertaining cases through outpatient clinics and hospitals, the populations of these studies differ. Los Angeles County excluded paediatric cases from their primary analysis, whereas almost half of our cases (and controls) were children. Because other studies have found associations between CA-MRSA and incarceration and drug use [Reference Miller26–Reference Hota28], participants in our study were questioned regarding incarceration and drug use, but no participants reported exposure to either. A case-control study conducted by Texas Children's Hospital in Houston comparing CA-MRSA to CA-MSSA infections did not find prior antimicrobial use to be associated with CA-MRSA; however, antimicrobial history was ascertained by parent/guardian interview only [Reference Sattler, Mason and Kaplan29]. Other studies with methods similar to those used here have observed associations between prior antimicrobial use and CA-MRSA infections compared to CA-MSSA infections [Reference Skiest30, Reference Davis31] and CA-MRSA infections to uninfected controls [Reference Schneider-Lindner32].
Although both CA-MRSA and CA-MSSA cases were more likely than uninfected community controls to be non-white and have lower household incomes, CA-MRSA cases were more likely to have these demographic features. The reasons for these differences are unknown but they have been demonstrated in previous studies [Reference David27, Reference Sattler, Mason and Kaplan29, Reference Skiest30, Reference Schneider-Lindner32].
Having a smoker in the home was significantly associated with CA-MRSA infection compared to uninfected CA-MRSA community controls in the multivariate model. Passive exposure to cigarette smoke has been observed as a risk factor for bacterial infections [Reference Nuorti33]. Maternal smoking has previously been identified as a risk factor for CA-MRSA colonization in patients in intensive-care nurseries [Reference Seybold34]. However, there are notable differences between our study population and intensive-care nursery patients and findings from one study may not be generalizable to the other. Other studies of S. aureus colonization have found higher colonization rates in participants who are smokers and participants passively exposed to cigarette smoke [Reference Bogaert35, Reference Melles36], thought to be due to changes in the respiratory mucosa. Having a household member who was a smoker rather than the participant's smoking habits may have been associated with CA-MRSA in our study because of the high proportion of participants who are young children and are therefore unlikely to be smokers themselves. The association between household smoking and CA-MRSA needs further study, particularly because we did not observe any association between CA-MSSA and household smoking.
We asked extensive questions about the healthcare exposures of study participants and their household members. After controlling for socio-economic variables, we did not find an association between healthcare exposures and CA-MRSA infection. Other variables found to be associated with CA-MRSA in a univariate analysis were not associated using a multivariate model.
It is notable that 69% of the strains associated with CA-MRSA cases in our study were USA300 and USA400, which contain PVL. Although the site of infection was matched, most of the MSSA strains we evaluated did not contain PVL. The specific role of PVL in S. aureus disease is currently an area of intensive research. Although PVL has a known association with skin abscesses [Reference Naimi17, Reference Issartel37, Reference Lina38], its role and the role of other staphylococcal toxins in the virulence of CA-MRSA strains such as USA300 is still being evaluated [Reference Li39]. Sixteen percent of isolates classified as CA-MRSA were USA100, which is typically associated with HA-MRSA [Reference Naimi17]. This highlights the difficulty of defining CA-MRSA by epidemiological criteria as CA-MRSA strains are now often identified in healthcare settings and healthcare-associated strains are increasingly identified in patients meeting CA-MRSA criteria [Reference Saiman12, Reference David40].
A self-reported history of skin problems was associated with both CA-MRSA and CA-MSSA infections compared to community controls. However, unlike other studies [Reference Schlievert41], we did not find specific skin problems associated with either CA-MRSA or CA-MSSA. This may be because cases reported their current infection as a history of skin problems or because our definition of skin problems lacked specificity.
Our study has several limitations. First, the study was designed to be hypothesis-generating, not hypothesis-testing. Numerous potential risk factors were tested and specific details were not obtained for each risk factor. For example, we did not collect the reason, dose or duration of antimicrobial prescribed or if the patient filled and completed the prescription. We did ascertain the type of the antimicrobial prescribed; however, our study did not have enough statistical power to determine if certain antimicrobial classes other than β-lactams were associated with CA-MRSA (e.g. macrolides) and if other classes were not. We also limited the analysis to antimicrobials used 1–6 months prior to culture, excluding those prescribed within 30 days of culture because antibiotics given at that time may have been used to treat the current staphylococcal infection. However, excluding that time period did not allow us to evaluate it. Because the study was intended to be hypothesis-generating and the sample size was relatively small, further study of risk factors found to be associated with CA-MRSA should be conducted. Most infections were SSTIs, and although the distribution of infection sites in our study is similar to that seen elsewhere for CA-MRSA [Reference Fridkin42, Reference Buckingham43], our results are not generalizable to cases with invasive infections.
Conclusions regarding differences between CA-MRSA and CA-MSSA and community controls may be limited. Persons with recent S. aureus infections may be more likely to remember previous skin problems or providers seen in the 6 months before their infection and this may have influenced results. Because fewer CA-MRSA and CA-MSSA community controls than cases permitted us to obtain antimicrobial history, antimicrobial use may be underreported in community controls. However, there were no significant differences in the proportion of CA-MRSA community controls and CA-MSSA community controls for whom we obtained antimicrobial history. We do not have information on patients with CA-MSSA infection who did not consent to have their records released to MDH or information on households who refused participation as community controls. Participation bias may have influenced our findings, especially with respect to associations between socio-economic factors and the risk of infection.
Over the past decade, CA-MRSA has evolved from an emerging infection to one that is commonly seen in all healthcare practice settings and in patients in all socio-economic strata. Although treatment options are available, there is evidence that resistance to additional antimicrobial classes is emerging [Reference Buckingham43–Reference Diep45]. If this trend continues, providing effective treatment will become increasingly difficult. Judicious antimicrobial use must continue to be emphasized in clinical care and clinicians as well as the public need ongoing education about the risks of inappropriate use of antimicrobials. In addition, continued emphasis on optimal hygiene and wound care to prevent transmission of all pathogens should be encouraged.
ACKNOWLEDGEMENTS
The study was supported by a cooperative agreement with the Centers for Disease Control and Prevention as part of the Emerging Infections Program, Grant no. 5 U01 CI000313-03. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry.
We acknowledge the following persons who contributed to the research and preparation of this article. Connecticut Department of Health: James Hadler, Minnesota Department of Health: Elizabeth Barksdale-Boyle, Joanne Bartkus, John Besser, Elizabeth Cebelinski, Richard Danila, Candace Fuller, Alanna Gramenz, Jessica Jungk, Billie Juni, Melissa Kemperman, Lindsey Lane, Christine Lees, Lindsey Lesher, Summer Martins, Jessica Nerby, Anne Potts, Tim Naimi, Beth Shade, Renae Waneka. Minnesota hospitals: Pat Ackerman, Dorothy Berg, Dina Bougie, Kathy Gray, Sally Petrowski, Michael Olesen, Adriene Thorton, Cindi Welch, and Ann Zierden.
DECLARATION OF INTEREST
None.








