Background
Foodborne outbreaks in England and Wales are notified to Public Health England (PHE) as part of a European Union directive (Directive 2003/99/EC [1]). The average annual number of outbreaks reported over the last 10 years (2008–2017) was 60 [2]. The majority of outbreaks that are reported are found to be caused by Salmonella, Campylobacter or Norovirus [Reference O'Brien3–Reference Adak5].
On the morning of Monday 4 March 2013, Newcastle City Council Environmental Health (EH) team began to receive reports of gastrointestinal illness from attendees at ‘Street Spice’, a food festival held between Thursday 28 February and Saturday 2 March 2013. The street food festival was advertised as ‘the North East's largest spice food festival’. The festival was held in a large temporary marquee in the city centre and included 25 stalls serving hot and cold food and drinks provided by different vendors from around the UK. The event was well-publicised, attracting visitors from across the North East of England. The organisers estimated that 10–12 000 people attended over the 3 days.
By Tuesday 5 March more than 20 reports had been received and a multi-agency outbreak control team (OCT) was convened to coordinate the epidemiological, microbiological, and environmental investigation to identify the source of infection, estimate the true burden of illness and implement control measures to prevent further cases.
Methods
Hypothesis generation
Cases who self-reported to the EH department were interviewed to collect information on exposures at the festival.
Epidemiological investigation
Symptomatic individuals were identified by a variety of methods including self-reporting, supplemented by alerts to general practitioners and hospital laboratories, messages posted on Twitter and Facebook pages hosted by the event and media statements about the outbreak.
Cohort study
A cohort study was undertaken using a web-based survey to collect food exposure and illness details (sent Friday 8 March). The survey was publicised using the event's social media pages which had a large following and through local newspapers and television coverage, requesting that all attendees participate, whether ill or not. Two sets of reminders were issued by social media.
The case definition was ‘a person who developed a diarrhoeal illness between 12 h and 5 days after attending the Street Spice festival’.
A household secondary case was defined as ‘a person who did not attend the Street Spice festival and developed diarrhoeal illness after the onset of a case within their household’.
We compared stalls visited (yes, no or don't know), foods eaten (yes, no or don't know) and days attended between cases and non-cases using univariable analyses to calculate odds ratios (OR). ORs were chosen as the measure of association as it was expected that the response rate would be low among attendees at the festival (approximately 12 000) and that there was likely to be a participation bias associated with illness (i.e. higher response rate in those developing illness) [Reference Charlett6]. We conducted a stratified analysis of the variables by the main risk factors identified. We used multivariable logistic regression to identify independent risk factors associated with illness. All risk factors that had a P value less than 0.2 and an OR greater than 1 were considered in a multivariable analysis. The final model included risk factors that were significantly associated with illness i.e. P value <0.05. Collinearity within the multivariable model was assessed. A sensitivity analysis was completed for the association to Stall A to assess participation bias using R [Reference Greenland7].
Follow-up study
During the course of the investigation, when it became apparent that some cases were reporting prolonged symptoms, the OCT agreed that a follow-up study of cases should be undertaken. Cases reported by any method were sent a link to the follow-up survey by email if available, otherwise by post. Cases who reported ongoing illness in the case follow-up study were re-contacted on 10 May to determine the duration of symptoms.
Capture re-capture study
A three-source capture-recapture analysis was undertaken to estimate the true number of people affected, using available sources of information on case numbers. The sources included illness self-reported to the EH department, laboratory results of people whose samples were submitted independently through routine health care services and people reporting illness via the web-based questionnaire for the cohort study. The individual datasets were de-duplicated and subsequently matched using exact matches followed by fuzzy matching, on the name, date of birth, gender and address variables, using Jaro Winlker [Reference Jaro8] and Damerau Levenshtein [Reference Oommen and Loke9] string distance algorithms. Resulting matches were manually assessed by the researchers. Log-linear Poisson regression analysis was applied to the final linked dataset [Reference Sandland and Cormack10–Reference Gerritse, van der Heijden and Bakker12].
Microbiological investigation (human samples)
Stool samples from symptomatic individuals were tested for Salmonella, Campylobacter, Escherichia coli O157, Shigella and Cryptosporidium using methods as described in UK Standards for Microbiology Investigations [13, 14]. In accordance with local outbreak investigation protocol, initial samples were also tested for Clostridium perfringens, Bacillus cereus and Norovirus. These additional tests were not undertaken on later samples as microbiological and clinical information did not indicate that these were likely causative agents. All Salmonella isolates were sent to the Gastrointestinal Bacterial Reference Unit laboratory, Public Health England (PHE; formerly the Health Protection Agency) for further sero- and phage typing.
As Salmonella was isolated in a relatively small proportion of faecal samples, additional investigations were undertaken. Specimens were re-cultured using local standard methods of Xylose Lysine Deoxycholate agar and selenite enrichment broth. Following overnight incubation, broths were inoculated onto Oxoid Brilliance™ Salmonella Agar in addition to bioMerieux chromogenic media. This did not increase the detection of Salmonella so all faecal specimens (including those both positive and negative for Salmonella by a culture where sufficient material remained) were tested at the PHE regional laboratory in London, using a multiplex polymerase chain reaction (PCR) for a range of gastrointestinal bacterial pathogens [Reference Moran-Gilad15]. The multiplex PCR included targets for Salmonella, Salmonella subspecies 1, Campylobacter (coli and jejuni), verocytotoxin-producing E. coli (VT 1 and VT 2), Shigella/entero-invasive E. coli (EIEC) and entero-aggregative E. coli (EAEC). Faecal specimens positive for EAEC were serotyped using antisera raised in rabbits to the E. coli somatic O antigens.
Additional univariable analyses were undertaken on cases where confirmed microbiological results were available. Multiple case definitions were used; one for each specific pathogen identified e.g. original case definition plus positive for EAEC. Each specific pathogen case definition was compared with non-cases from the original cohort study i.e. non-symptomatic individuals.
Food and environmental samples were analysed at the PHE Food, Water & Environmental Microbiology Laboratory for E. coli and enterobacteriaceae, as a marker of faecal contamination and the following specific pathogens; Salmonella, L. monocytogenes, C. perfringens, Bacillus spp., E. coli O157 and S. aureus.
Environmental investigation
Information was gathered about all the stalls at the event. Early case interviews identified that all people reporting illness had eaten food from an individual stall, hereafter referred to as Stall A and further investigations were concentrated on Stall A.
Detailed information about food preparation including descriptions of ingredient handling and the sources of each ingredient were collected.
No prepared food from the event remained, so samples from the same batches of a number of different ingredients used were collected. Environmental samples were not collected from areas where food was served as stalls had been dismantled by the time illness was reported. Environmental samples were collected from the stall holder's permanent restaurant kitchen where the majority of food from Stall A was initially prepared during the event, before being transported to the marquee where it was further stored and served.
Results
Hypothesis generation
Information from interviews with the first reported cases suggested that consumption of food from an individual stall, hereafter referred to as Stall A, was a common exposure.
Descriptive epidemiology
The total number of people reporting illness (via EH Officers (EHO), laboratories and the web-based survey) after attending the festival was 592; 263 males (44%) and 311 females (53%), 18 had unknown gender. The median age was 34-years-old (range 0 − 100 years).
The capture-recapture analysis estimated that the total number of people ill was 926 (95% CI: 628–1224).
There were 827 responses to the web-based cohort study survey; there was no significant difference in age or sex between responders and non-responders of the survey for cases. Of the respondents, 347 reported gastrointestinal symptoms following the event of which 306 that met the case definition. The mean incubation period for the cases was 1.5 days (range 1–4 days) (Fig. 1).
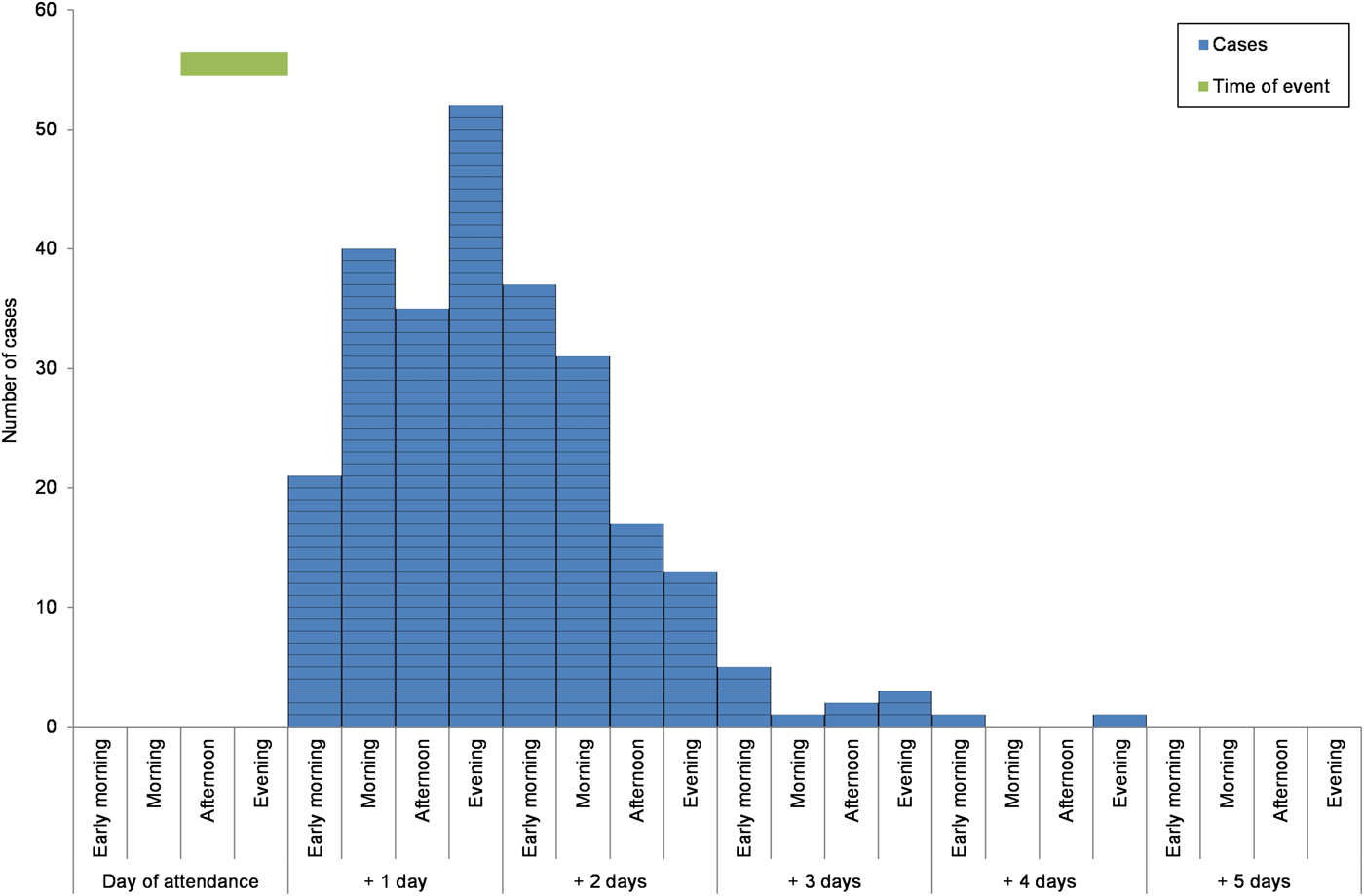
Fig. 1. Epidemic curve of onset by ‘days after attendance at the event’ for the cases with gastrointestinal disease that participated in the cohort study, North East England, March 2013.
Of the 592 cases reported by any method, 527 were sent the case follow-up survey and 374 responded (71%). Of these 351 met the case definition (94%). Those not meeting the definition were cases reported to EHOs but who had not completed the web-based questionnaire. Symptoms were predominantly diarrhoea (100%) and abdominal pain (91%). The median duration of symptoms was 11 days (range 1–49 days) for diarrhoea and 7 days (range 1–34) for abdominal pain. Seventy percent of cases who were employed took time off work (207/296). The mean number of work days missed was 5 and the range of days off work was 1–35. The total number of days off work recorded for study respondents was 995 days (N = 202, 5 cases who missed work did not provide a number of days missed). Two cases were hospitalised (0.5%).
Analytical studies
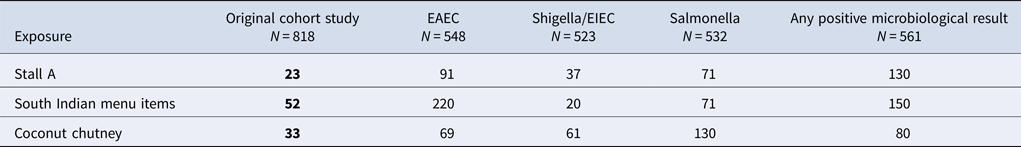
Univariate analysis from the cohort study found a significant association between illness and eating food from the Stall A (OR = 23; 95% CI 15–37) and eating the South Indian foods (OR = 52; 95% CI 33–83). 80% of the cases reported eating South Indian foods (Table 1). There were no statistically significant positive associations between being ill and any of the other stalls. Participation bias was assessed for Stall A and the result was found to be robust; even with a low response from non-cases and those not exposed (25%), the OR remained significant (original OR: 23, bias-corrected OR: 54).
Table 1. Univariable analyses of the selected stall and food exposures from the event and their association with illness from the cohort study of attendees of the food festival, North East England, March 2013

*Total number of stalls assessed was 20; total number of food items assessed was 16.
When specific food items served at Stall A were considered, significant associations between illness and eating some food items were observed (Table 1). These included consumption of all main dishes; dosa (OR = 24; 95% CI 15–37), uttapam (OR = 30; 95% CI 7.4–260) and vada (OR = 14; 95% CI 6.3–33), all of which were served with coconut chutney as an accompaniment. Consumption of coconut chutney was associated with the greatest odds of illness (OR = 33; 95% CI 20–57).
When stratified by consumption of foods prepared by the guest chef (South Indian foods), associations between illness and consumption of standard restaurant foods consumed from Stall A (i.e. usual chef or non-South Indian foods) were reduced (univariable OR 2.4, adjusted OR 2.2; −10% change) suggesting that confounding between the different foods had raised the univariable ORs. When individual Stall A food items were stratified by coconut chutney associations of other food items were similarly reduced (average decrease of 75% in adjusted OR).
The final multivariable model included Stall A (aOR 3.1, 95% CI 1.8–5.5), any South Indian food (aOR 16, 95% CI 7.8–31) and coconut chutney (aOR 2.1, 95% CI 1.0–4.3). Having any South Indian food had the highest odds of being associated with illness.
Microbiological results
Of the 110 faecal specimens submitted to local laboratories, including the North East HPA laboratory for testing, 29 cases of laboratory-confirmed Salmonella infection were detected, 25 of which were identified as Salmonella Agona phage type (PT) 40, a strain not previously seen in human or food isolates in the UK. Other serovars identified were S. Hadar, S. Cero and S. Typhimurium, the remaining isolate was untyped.
The molecular (multiplex PCR) testing performed on the subset of 77 case samples identified EAEC in 82% (n = 63) of the samples tested [Reference Moran-Gilad15], Shigella/EIEC in 42% (n = 32) and E. Coli VT1 in 8% (n = 6). Multiple pathogens were detected in 42% (n = 32) of this subset of samples (Table 2). Although PCR multiplex assay is not a standard clinical diagnostic technique, detection of multiple gastrointestinal pathogens was consistent with the prolonged and severe diarrhoea reported by cases.
Table 2. Number of cases with multiple pathogens detected by PCR and culture (N = 32), details of symptom composition (%), duration of diarrhoea and incubation period (median days)

a These fields were only available for those that completed the follow-up survey (N = 53, 69% of those with PCR results).
b D = Diarrhoea, A = Abdominal pain, V = Vomiting and F = Self-reported fever.
c ‘–’ indicates that there were no cases or no information available for the cases in that category.
The EAEC isolates were assessed further to identify serotypes; the most commonly identified were O131:H7 (6 isolates) and O104:H4 (5 isolates). None of the EAEC O104:H4 isolates in this study had the stx gene but it was found that they share a common ancestor with the German outbreak strain from 2011 [Reference Dallman16, Reference Grad17]. Salmonella Agona PT 40 was isolated from fresh uncooked curry leaves collected from the same batch used in coconut chutney served at the event. Pulsed-field gel electrophoresis showed that human and food S. Agona isolates were indistinguishable (Fig. 2).
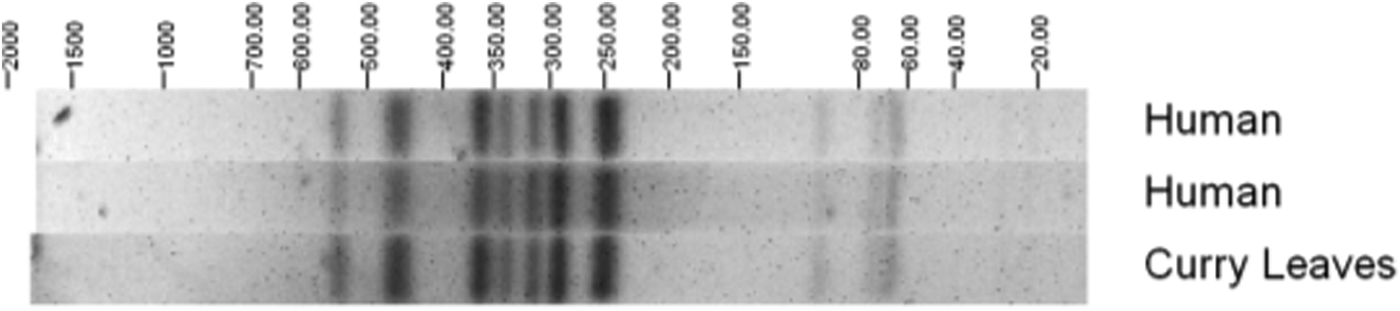
Fig. 2. Pulsed-field gel electrophoresis profiles of Salmonella Agona PT 40 isolates from cases and a curry leaf sample.
E. coli detected in food samples, including curry leaves, was identified by standard culture methods. It is not standard practice to undertake further typing of E. coli identified in food specimens. The standard culture method would identify EAEC as E. coli but would not differentiate further.
In univariable analyses using the different pathogen case definitions, exposures to the same stall and food items as found in the cohort study using the symptomatic case definition were associated with illness at a greater magnitude (Table 3).
Table 3. Odds ratios from univariable analyses comparing selected risk factors against different laboratory confirmed pathogen case definitions from the cohort study of the attendees of the food festival, North East England, March 2013
Environmental investigation
Here we report solely the results for Stall A.
The majority of food from Stall A was prepared in the stall holder's permanent restaurant kitchen and transported in covered containers to the venue (travel time less than 15 min) Stall A had two sections, both of which served Indian food. One section served food from the stall holder's usual restaurant menu and the other section had South Indian food prepared by a guest chef.
The South Indian dishes were not included in the usual restaurant menu. The stall holder estimated that 900–1000 portions of the guest chef's South Indian food were served over the three days of the festival. The South Indian food served at Stall A were vegetarian pancake-type foods (dosa, uttapam and vada). All were served with coconut chutney and lentil sambar sauce unless requested otherwise. The batter for the dosa and uttapam was made by soaking dried lentils and rice overnight before blending the following day. The dosa filling of vegetables, spices and curry leaves and the vada (deep-fried chickpeas, spinach and spices) were cooked and chilled in advance. Dosa filling was reheated and hot-held at the venue and vada were reheated to order. Uttapam topping (onions, pepper and coriander) was cooked to order at the venue.
Lentil sambar (slow-cooked lentils, onions, tomatoes and spices) was cooked and chilled in advance and reheated and hot-held at the venue. Coconut chutney was prepared in advance in a designated area of the kitchen. Coconut milk, desiccated coconut, ginger, chillies, coriander and curry leaves were blended with water and seasoning and subsequently served uncooked. Several bags of fresh uncooked curry leaves were purchased from a local supplier on 27 February and leaves were stripped from their stems and refrigerated together in one bag. The curry leaves were washed under running cold water before being added to other ingredients and were not in contact with any other kitchen surfaces. Batches of chutney were made on each day of the event, refrigerated in covered containers and transported to the venue in the morning. A large refrigerator was present at the venue for storage of pre-prepared foods. However, a small serving bowl of coconut chutney held at ambient temperature was present on Stall A.
Food safety and control measures
During the course of the investigation, the OCT identified three key areas of potential public health risk which were investigated and control measures were implemented in a timely fashion.
(1) A contaminated food item remaining in the food chain with the risk of further cases. The use of fresh uncooked curry leaves was not the usual practice in the restaurant/stall implicated in this outbreak. Consequently, the OCT did not consider there to be an on-going risk to local public health associated with this incident. However, the OCT identified a potential public health risk with the use of fresh uncooked curry leaves by other food businesses in the UK.
(2) An infected food handler as the source of infection. In order to investigate this, all staff involved in preparing and serving food were interviewed and submitted stool specimens for routine testing. None were symptomatic and all samples were negative for Salmonella. Samples were not tested for EAEC.
(3) Secondary transmission of infection by cases. All cases were given verbal hygiene advice and recommended to remain absent from work until 48 h symptom-free [18]. Seventeen household secondary cases were indicated in the case follow up survey (no further details were collected).
The supply chain of fresh curry leaves used in this incident was investigated, including sampling at the point of entry to the UK. Curry leaves were produced in Pakistan and washed before packaging. Microbiological testing of the leaves was not undertaken by the producer but results of microbiological testing of washing water were available. Curry leaves were received in the UK as a wrapped product and no further packaging or labelling was undertaken. Salmonella was not detected by the PHE FWE laboratory in samples collected after the outbreak (samples collected in April 2013). Additionally, to mitigate the wider public health risk the OCT worked closely with the Food Standards Agency to consider control measures, including a sampling survey of imported fresh curry leaves and specific advice to food business operators.
In accordance with requirements and to enable any linked cases detected in Europe to be considered, an alert was sent to the European Centre for Disease Control (ECDC) Epidemic Intelligence Information System (EPIS) on 19 March 2013. No reports were received from European partner agencies of further cases of S. Agona PT40 or illness associated with attendance at the event.
Discussion and conclusions
We describe the investigation of one of the largest outbreaks of gastrointestinal illness in England, which included cases of laboratory-confirmed S. Agona PT 40, which had not previously been detected in the UK and multiple other pathogens. This outbreak was caused by the use of fresh uncooked curry leaves which were found to be contaminated with the same strain of S. Agona PT 40 detected in faecal samples and also potentially with other gastrointestinal pathogens. A review of outbreaks reported to the PHE's electronic Foodborne and non-foodborne gastrointestinal Outbreak Surveillance System (eFOSS) [19] found that this was one of the largest food-borne outbreaks in the UK since 1992 and was the largest UK outbreak where herbs or spices had been identified as the suspected source of infection.
Microbiological findings from human and food samples were supported by epidemiological evidence from a large cohort study which identified that coconut chutney made with fresh uncooked curry leaves had the strongest univariable association with illness. The multivariable association with coconut chutney may be lower than expected due to either misclassification, as the accompaniment was likely to have been added to all dishes without the knowledge of the attendee, or because other foods from the implicated stall also acted as vehicles through cross-contamination from the suspect dish.
The use of molecular (multiplex PCR) testing in a gastrointestinal outbreak investigation was a novel approach in 2013. It was undertaken on a subset of faecal samples to explore the disproportionately large burden of illness compared with the relatively low proportion of tested cases with laboratory detection of Salmonella by routine culture methods. The multiplex GI PCR used was developed by PHE for rapid testing during the 2012 Olympics and clinical validation showed good sensitivity and specificity for a majority of clinical targets [Reference Severi20, 21]. Multiple additional pathogens including EAEC and Shigella/EIEC were detected by PCR but not by traditional culture methods.
Reported symptoms of cases without Salmonella detection were consistent with those of EAEC and Shigella infection. EAEC is a common cause of gastrointestinal illness in travellers to developing countries [Reference Dallman16, Reference Adachi22], typically causing abdominal pain and diarrhoea that may persist for more than 2 weeks [Reference Nataro, Steiner and Guerrant23]. EAEC has been associated with illness globally [Reference Nataro, Steiner and Guerrant23] but is often undiagnosed [Reference Huang24, Reference Chattaway25] and outbreaks have been reported around the world [Reference Chattaway25] including the UK [Reference Smith, Cheasty and Rowe26]. Similarly, Shigella is endemic in Asia, the source of the contaminated curry leaves and may cause persistent diarrhea [Reference Ram27].
Infection with multiple pathogens was detected in nearly one-third of the subset of samples submitted for PCR testing. Co-infection with different gastrointestinal pathogens has been described in the UK in the IID2 study but was uncommon and more frequently involved viruses [Reference Tam28]. However, mixed infections are likely to be detected more frequently as microbiological tests improve [Reference Chattaway25]. Mixed infections may reflect common exposure such as sewage contamination of water with multiple pathogens [Reference Tam28] and the organisms detected in this outbreak were consistent with those likely to be found in sewage-contaminated water.
Mixed pathogen outbreaks involving co-infection with bacterial pathogens have been reported infrequently and may be under-reported [Reference Berkelman29–Reference Wensley and Coole32]. It has been suggested that microbiological investigation of an outbreak may stop once a pathogen is detected [Reference Layton30]. Consequently, while the detection of alternative or multiple pathogens is dependent on available routine diagnostic tests this also requires the outbreak investigation team to remain mindful of opportunities for additional testing. It will be important for investigating teams to share findings on the clinical significance of mixed pathogen outbreaks to increase the knowledgebase and help to plan investigations accordingly in the future.
Limitations of our investigation were that, in accordance with standard laboratory methods [33] for investigation of food samples, further characterisation of E.coli was not undertaken. We were, therefore, unable to determine whether curry leaves were contaminated with EAEC. Additionally, although validated for most targets, the GI PCR was not a routine clinical test. However, we believe that these results show that a range of faecal organisms contributed to the burden of illness, with the most likely source being contaminated curry leaves.
Potential contamination of fresh and dried herbs with pathogenic organisms including Salmonella is well-documented in the scientific literature and outbreaks have been reported [Reference Elvis34, Reference Zweifel and Stephan35]. A sampling survey conducted by the PHE detected Salmonella (S. Newport, S. Virchow PT 8 and S. Virchow PT 21) in 2/21 samples of curry leaves, although none of the leaves tested in that survey were from Pakistan [Reference Dallman16]. It is, however, unclear whether there is a widespread understanding among food handlers and the public about this potential risk.
Fresh herbs are commonly used in commercial and domestic settings and may be consumed raw or added to food after cooking depending on local culinary practices. Used in this way, herbs are considered to be in a ready-to-eat state, without the need for cooking or other processing effective to eliminate or reduce micro-organisms of concern to an acceptable level [36]. Information gathered during our investigation indicated that food handlers did not consider the use of fresh uncooked curry leaves to be associated with significant risk of food poisoning. The outbreak report prompted media comments by several famous chefs not associated with this incident affirming the safety of fresh uncooked curry leaves and questioning the possibility of plants being a source of Salmonella. As noted, although the exporter considered this product to be raw, this was not included in labelling and those purchasing these leaves may have been unsure whether they were safe to use without further processing. At the time of the outbreak, there was no published guidance from the UK Food Standards Agency regarding the use of curry leaves and their status as a ready to eat product
Plants, including herbs, may be exposed to contaminated water either during irrigation or post-harvest processing [Reference Berger37]. Curry leaves were reported to have been washed in Pakistan prior to export and were washed in the restaurant prior to use. However, the attachment mechanisms exhibited by EAEC enable the bacteria to adhere tightly to leaves [Reference Berger38] and washing may have been insufficient to remove all pathogens [Reference Berger37].
Our investigation identified that the use of fresh uncooked curry leaves in a finished dish was not the usual practice at the restaurant managing Stall A. A change to usual practice without an associated risk assessment has been implicated as a factor in other reported outbreaks [Reference Wensley and Coole32]. The OCT tried to establish whether the use of fresh uncooked curry leaves was considered ‘normal practice’ when preparing coconut chutney and several different recipes were identified including both raw and ‘tempered’ leaves (where leaves are briefly heated). It was, therefore, not possible to establish whether the use of raw leaves is ‘accepted practice’ in this style of cooking.
The burden of illness associated with outbreaks is difficult to quantify and, other than severity of illness and numbers hospitalised may not be included as part of a routine outbreak investigation. We used a number of different epidemiological methods including a capture-recapture analysis and a follow-up study to try and quantify the true burden of illness associated with this outbreak. These findings described a significant burden of disease; 70% of employed cases who responded to the follow-up study missed an average of 5 working days and we estimated the total number of cases could have been more than 60% greater than the number of individuals identified (926 vs. 592).
The investigation of this large outbreak highlighted a number of factors that public health teams should consider including the scale of the event, the introduction of unfamiliar food preparation processes and the uncertainty and conflicting views regarding the use of some food items. Our findings emphasise the importance of continuing to search for an infectious cause of illness when the clinical presentation of cases is not consistent with results from standard laboratory tests. The detection of previously underreported organisms and mixed pathogen outbreaks may increase with improvements in diagnostic methods and the more widespread use of molecular testing for gastrointestinal pathogens.
Acknowledgements
The authors would like to thank all members of the outbreak control team, environmental health officers and teams within local laboratories for their contributions to this outbreak. The authors would also like to thank the participants of the various studies.
Financial support
None
Conflict of interest
None








