Evidence from animal studies supports a causal link between low-grade inflammation, insulin resistance and impaired intestinal barrier function( Reference Cani, Amar and Iglesias 1 , Reference Cani, Bibiloni and Knauf 2 ); however, we recently demonstrated for the first time that intestinal permeability (IP) is compromised in type 2 diabetes (T2D) patients compared with healthy age- and BMI-matched volunteers( Reference Horton, Wright and Smith 3 ). Increased small IP as measured by urinary excretion of orally administered 51Cr-EDTA was significantly and positively correlated with the inflammatory marker TNF-α. This may indicate that the chronic systemic low-grade inflammation characterising metabolic diseases such as T2D is associated with a leaky gut in humans.
It is hypothesised that the impaired intestinal barrier leads to an increased translocation of the gram-negative bacteria cell membrane component lipopolysaccharide (LPS) (as well as whole bacteria and other luminal antigens) into the circulation, which results in metabolic endotoxaemia. LPS is a ligand of the toll-like receptor 4 (TLR-4). Activation of TLR-4 signalling by LPS results in a low-grade inflammation, which affects insulin signalling and thus induces insulin resistance( Reference Cani, Amar and Iglesias 1 ). Interestingly circulating LPS is indeed elevated in T2D compared with healthy controls( Reference Creely, McTernan and Kusminski 4 , Reference Pussinen, Havulinna and Lehto 5 ). However, whether this is due to increased paracellular movement or due to fat-induced LPS absorption through increased chylomicron formation is unclear( Reference Ghoshal, Witta and Zhong 6 ).
Intestinal dysbiosis in T2D has been observed in a number of cross-sectional studies( Reference Larsen, Vogensen and van den Berg 7 – Reference Lambeth, Carson and Lowe 12 ). Larsen et al.( Reference Larsen, Vogensen and van den Berg 7 ) found that Betaproteobacteria and the Bacteroidetes:Firmicutes ratio correlated positively with plasma glucose concentrations. Thus, as a potential therapeutic target, altering intestinal bacterial community structure and thereby reducing LPS load and uptake, may be beneficial in T2D. An approach to changing the intestinal bacterial composition by diet is with the use of prebiotics and probiotics. Studies in rodents suggest that prebiotics, probiotics and synbiotics may improve intestinal barrier function and glucose control( Reference Cani, Bibiloni and Knauf 2 , Reference Mattace Raso, Simeoli and Iacono 13 – Reference Everard, Belzer and Geurts 15 ). However, few studies have investigated the use of prebiotic supplementation in human T2D( Reference Pourghassem Gargari, Dehghan and Aliasgharzadeh 16 – Reference Dehghan, Farhangi and Tavakoli 22 ) and none in terms of the potential mechanistic effects on the intestinal barrier. This is the first study to investigate the effects of prebiotic supplementation on intestinal bacteria, IP, endotoxaemia and glucose tolerance concurrently in T2D patients.
Methods
This was a randomised, double-blind, placebo-controlled parallel study comparing the effects of prebiotic supplementation with placebo treatment for 12 weeks on glucose control, IP, intestinal bacterial composition, endotoxaemia and inflammatory markers in patients with T2D. The protocol was approved by the Central London NRES Committee (REC reference no. 11/LO/1141) and the University of Surrey Ethics Committee and was conducted according to the declaration of Helsinki. The trial was registered at the UK Clinical Research Network portfolio database under trial identifier ISRCTN07813749.
Subjects
Men with well-controlled T2D aged 42–65 years were recruited through local general practices and advertisement in a local newspaper. Because of the repeated administration of the radioactive compound 51Cr-EDTA and the potential influence of the menstrual cycle on outcomes, women were excluded from the study. All patients provided written informed consent. Exclusion criteria included the use of antibiotics in the past 3 months, use of anti-inflammatory medications (except a low-dose (75 mg/d) aspirin), diuretics, proton-pump inhibitors, inflammatory bowel disease, Crohn’s disease, coeliac disease and irritable bowel syndrome. Patients were asked to exclude probiotic products and prebiotic supplements (other than the study supplement) from their diet for 2 weeks before the first study visit and throughout the study. Furthermore, they were asked not to change their lifestyle during the study. The sample size for this study was based on the primary outcome measure of changes to IP and based on our own published pilot data using this method in patients with well-controlled T2D( Reference Horton, Wright and Smith 3 ). In this parallel-design study, thirty patients provided 80 % power to detect a treatment difference between groups of 1·6 % in total permeability, using the calculated sd in this cohort of 1·57 (α 0·05).
Study protocol
After the screening procedure, patients were randomised to either prebiotic fibre (galacto-oligosaccharide mixture (GOS mixture); Bi2muno) or placebo (maltodextrin) supplementation for 12 weeks according to a randomisation scheme generated at randomization.com. Both supplements were supplied by Clasado Ltd as dry white powders in sachets each containing 5·5 g and were readily mixed into beverages or food. The GOS mixture has been used in previous trials and is described by Vulevic et al.( Reference Vulevic, Juric and Walton 23 ). A dose of 5·5 g of GOS mixture has previously been demonstrated to have a bifidogenic effect in healthy individuals of this age and BMI, and be well tolerated in terms of gastrointestinal effects( Reference Vulevic, Juric and Walton 23 , Reference Davis, Martinez and Walter 24 ). Patients were contacted twice during the 12-week supplementation to monitor side effects and compliance. Patients returned unused sachets following the supplementation to verify compliance. Dietary intake data (7-d diet diary), clinical data and faecal samples were collected at baseline and at the end of the intervention. The diet diaries were analysed in DietPlan6 (Forestfield Software Ltd). Faecal samples were collected in sterile universal polystyrene containers and were kept refrigerated. Faecal samples were stored at −20°C initially and in a −80°C freezer for long-term storage.
The co-primary outcomes of the study were changes in IP, endotoxaemia and glucose tolerance. Secondary outcomes were changes in intestinal bacterial composition, inflammatory markers, lipids, blood pressure and anthropometric measurements. Use of metformin was considered a confounding factor. However, as thirteen out of fourteen patients in the prebiotic group were metformin treated, it was not possible to perform a subgroup analysis to explore a potential interaction between metformin and prebiotic treatment.
Intestinal permeability
IP was measured by 24-h urinary excretion of orally administered 51Cr-EDTA, as previously described( Reference Horton, Wright and Smith 3 ). We utilised 51Cr-EDTA as a probe, as it is stable in the colonic luminal environment allowing assessment of colonic permeability and it is easily detected in the urine( Reference Teshima and Meddings 25 ).
Anthropometric and blood pressure measurements
Having fasted overnight, patients attended the CEDAR Centre of the Royal Surrey County Hospital. Body weight and body composition were measured by bioimpedance (Tanita). Waist circumference was measured at the level of the navel with a tape measure. Blood pressure was measured on the non-dominant arm after 5 min of rest in a semi-upright position, and the mean of three readings was calculated (Omron MX3 Plus; Omron Healthcare Europe).
Glucose tolerance, inflammatory markers and lipids
Glucose tolerance was assessed using a frequently sampled insulin-modified intravenous glucose tolerance test (IVGTT), as previously described( Reference Bodinham, Smith and Wright 26 ). Blood was collected in EDTA tubes for glucose, insulin and C-peptide and HbA1c measurements and into serum tubes containing clotting activator or pyrogen-free tubes for measurements of inflammatory markers, lipids and LPS in serum. Aprotinin was added to blood samples (200 kallikrein inhibiting units/ml blood) collected for C-peptide measurement. Blood samples were centrifuged at 3000 g at 4°C for 10 min, and serum and plasma were stored at −20 or −80°C.
Biochemical analyses
Whole-blood glucose concentrations were measured on an YSI 2300 STAT Plus™ (YSI Life Sciences) with an average intra-assay CV of 4·8 % and inter-assay CV of 5·8 %. Plasma insulin and C-peptide were analysed in duplicate using RIA (Millipore) with average intra-assay CV of 7·7 and 4·2 % and inter-assay CV of 12·6 and 6·4 %, respectively. HbA1c and serum high-sensitivity C-reactive protein were measured by the Surrey Pathology Partnership, an accredited laboratory, and serum IL-6 and TNF-α were measured using a Luminex platform and Biorad bio-plex kits and software (Bio-Rad). Serum TAG, total cholesterol, HDL-cholesterol and NEFA were measured on an ILab650 using commercially available kits (Randox Laboratories and Instrumentation Laboratory). All intra-assay CV were <2 % and inter-assay CV≤3 % for lipid measurements. LDL-cholesterol concentration was calculated using the Friedewald formula( Reference Friedewald, Levy and Fredrickson 27 ). LPS was measured in duplicate using Endosafe-MCS (Charles River Laboratories), as previously described( Reference Everard, Belzer and Geurts 15 ). Serum LPS-binding protein (LBP) and sCD14 concentrations were measured using commercially available kits according to the manufacturer’s instructions (Hycult Biotechnology). The average intra-assay CV were 3·9 and 8·5 % for LBP and sCD14, respectively.
Amplification and high-throughput sequencing
Amplification and sequencing were performed as previously described by Ellis et al.( Reference Ellis, Bruce and Jenkins 28 ). Further details are provided in the online Supplementary material.
Bioinformatics
The sequences were processed in Qiime( Reference Caporaso, Kuczynski and Stombaugh 29 ) using the AmpliconNoise( Reference Quince, Lanzen and Davenport 30 ) pipeline that utilises flowgram information of the sequences to correct for errors. The samples were demultiplexed by exact matching of both barcode and primer, and the sequences were filtered and trimmed on the basis of identification of low-quality signals( Reference Quince, Lanzén and Curtis 31 ). The filtered flowgrams were clustered to remove platform-specific errors and converted into sequences using the PyroNoise algorithm. The sequences had barcodes and degenerate primers removed before trimming at 400 bp. They were then further clustered by SeqNoise to remove PCR single-base errors. In the final step, the Perseus algorithm was used to identify chimeras.
The denoised sequences were classified using the standalone Ribosomal Database Project (RDP) classifier( Reference Wang, Garrity and Tiedje 32 ). From this, taxa frequencies at five different levels – phylum, class, order, family and genus – were calculated. In addition, a non-supervised approach was used; operational taxonomic units (OTU) were generated at 3 % divergence following pair-wise global sequence alignment and hierarchical clustering with an average linkage algorithm. After generating the abundance tables, multivariate statistical analyses in the context of metadata were done in R using Vegan package (http://cran.r-project.org/web/packages/vegan/) for obtaining α- and β-diversity estimates, as well as permutation ANOVA using distance measures (adonis function). For calculating α-diversity measures, the samples were rarefied to the minimum sample size, whereas for other statistics we log-normalised the abundance tables. Where appropriate, P-values were adjusted using the Benjamini–Hochberg method to control the false discovery rate.
Quantification of bacterial groups by quantitative PCR
Total bacteria, Bifidobacterium, Roseburia, Lactobacillus, Enterobacteriaceae, Clostridium leptum and Clostridium coccoides groups were quantified using quantitative real-time PCR (qPCR). The qPCR methods are described in the online Supplementary material.
Statistical analysis
Clinical outcomes and diet data are presented as mean values with their standard errors or medians and interquartile ranges as appropriate. Baseline values between groups were compared using an unpaired t test or Mann–Whitney test and within-group changes with a paired t test or Wilcoxon’s matched pairs signed-rank test as appropriate. Treatment effects were assessed by comparing differences in changes from baseline between groups using ANCOVA with baseline values as covariates or the Mann–Whitney test if log transformation did not normalise data distribution. AUC for glucose, insulin and C-peptide was calculated using the trapezoid rule. Glucose and insulin data were modelled using Bergman’s minimal model (MINMOD Millennium version), as previously described( Reference Bodinham, Smith and Wright 26 ). Homoeostasis model assessment (HOMA) for insulin sensitivity (%S), β-cell function (%B) and insulin resistance (IR) were calculated using the HOMA2 Calculator (http://www.dtu.ox.ac.uk/). Associations between changes in gut bacteria abundance, diet and clinical outcomes were assessed by Kendall’s rank correlations. Analysis of qPCR data were performed on log10 transformed values. The level of significance was set at P<0·05. Data were analysed using GraphPad Prism 6, SPSS versions 21 and 22 and R.
Results
Fig. 1 shows the flow chart for the study. Of the thirty-two patients recruited, two patients withdrew from the study because of gastrointestinal upset (n 1) and antibiotic treatment (n 1). Another participant in the prebiotic group was excluded from the data analysis because of antibiotic treatment. Characteristics of the twenty-nine patients who were included in the final data analyses are shown in Table 1. All patients had been on a stable treatment for at least 3 months before taking part in the study and had no changes to their medications during the study. Two patients in the placebo group did not undergo a full post-supplementation IVGTT because of venous access problems; however, a fasting blood sample was obtained from one of the patients and data from the initial 20 min of the IVGTT for the second patient were included in the data analysis.
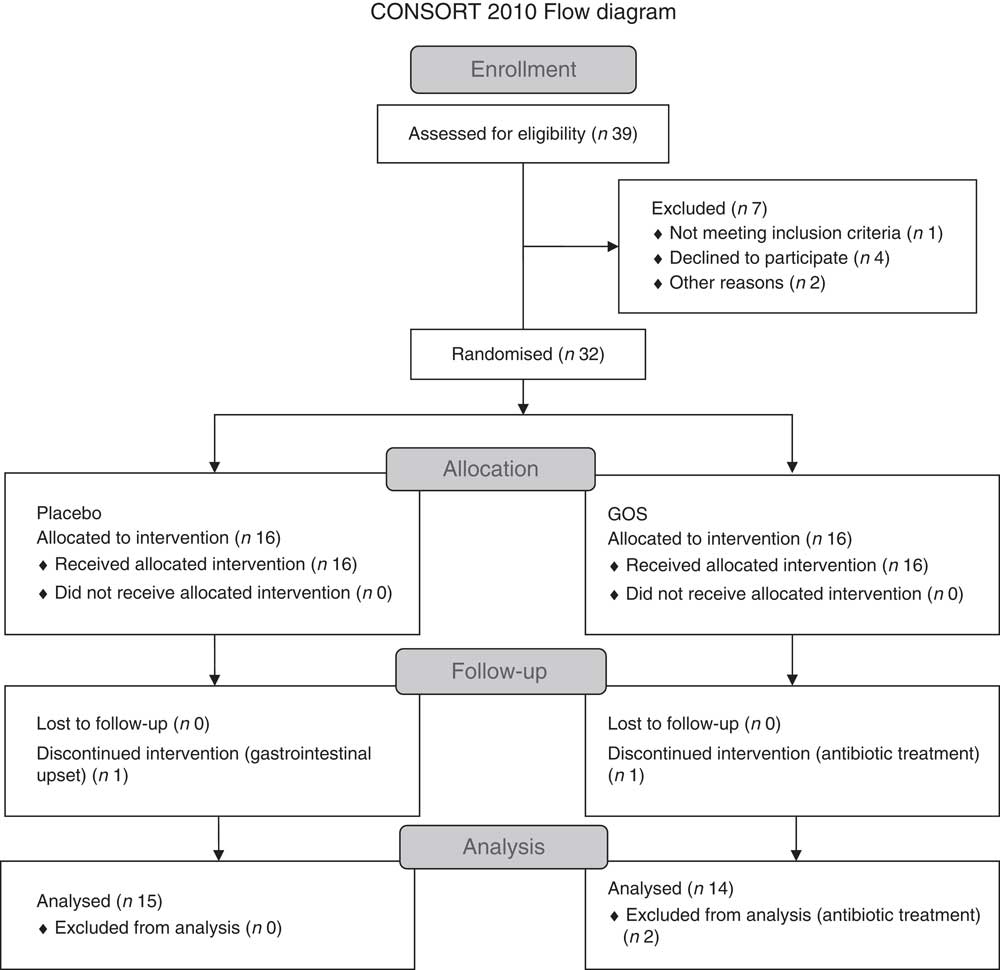
Fig. 1 Flow chart showing the recruitment and retention of patients in the study. GOS, galacto-oligosaccharide.
Table 1 Characteristics of the treatment groups at baseline (pre) and after supplementation (post) and diabetes medicationsFootnote * (Mean values with their standard errors; n 14 in the prebiotic group and n 15 in the placebo group)

* There were no differences in baseline (Pre) values between groups (P>0·05, unpaired t test).
† The P-value is for the comparison of the change between groups with Pre value as covariate (ANCOVA). Other medications (n) used by patients in the prebiotic group were statins (11), blood pressure medication (8), fenofibrate (2), omeprazole (2), low-dose aspirin (1), levothyroxine sodium (1) and citalopram (1). Other medications used in the placebo group were statins (8), blood pressure medication (8), low-dose aspirin (5), Omeprazole (2), benign prostate hyperplasia medications (2), hay fever medication (2), betahistine hydrochloride (1), asthma medication (1), medications for incontinence (2), sleep medication (1) and anti-fungal medication (1).
‡ n 13 in the placebo group.
§ Significant within-group change (P<0·05, paired t test).
|| n 13 in the prebiotic group.
¶ The remaining six patients in the placebo group were diet/exercise controlled.
Compliance, assessed by the number of unused sachets of supplement, was 96 % (range: 84–100 %) for both treatments. No adverse side effects were reported by the participants. There were no significant differences between groups in clinical outcomes at baseline; however, Enterobacteriaceae were higher (P=0·0379) (online Supplementary Fig. S2(e)) and Peptostreptococcaceae levels lower (P=0·0019) in the prebiotic group at baseline.
Anthropometrics and blood pressure
Supplementation with the prebiotic fibre had no significant effects on body weight, BMI, body fat percentage, waist circumference or blood pressure when compared with placebo (Table 1).
Intestinal permeability
Prebiotic supplementation had no significant effect on IP, as measured by urinary recovery of 51Cr-EDTA when compared with placebo (Fig. 2).
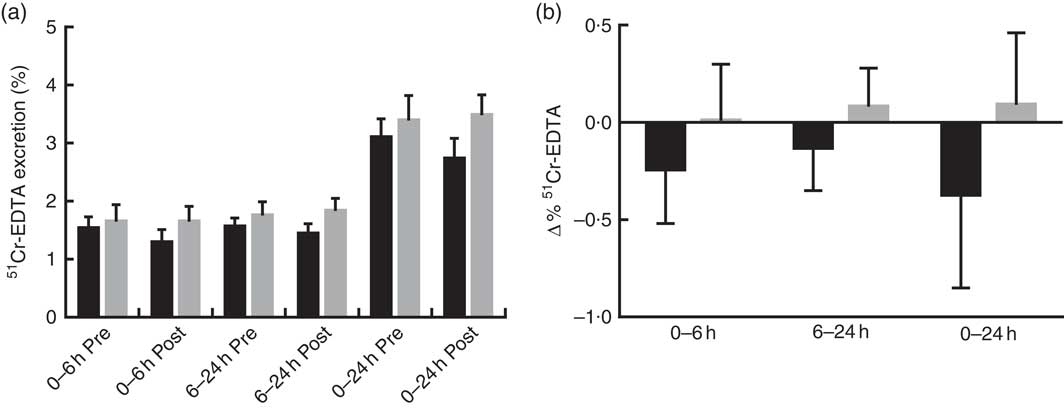
Fig. 2 Intestinal permeability estimated by 51Cr-EDTA excreted in urine following 12 weeks of prebiotic (![]() , n 14) or placebo (
, n 14) or placebo (![]() , n 15) supplementation. (a) Percentage 51Cr-EDTA excreted before (pre) and after supplementation (post) and (b) change in 51Cr-EDTA excreted. Values are means with their standard errors represented by vertical bars. There were no significant differences between treatment groups (P=0·322, P=0·235 and P=0·176 (ANCOVA) for small intestinal (0–6 h), colon (6–24 h) and total tract (0–24 h) permeability, respectively).
, n 15) supplementation. (a) Percentage 51Cr-EDTA excreted before (pre) and after supplementation (post) and (b) change in 51Cr-EDTA excreted. Values are means with their standard errors represented by vertical bars. There were no significant differences between treatment groups (P=0·322, P=0·235 and P=0·176 (ANCOVA) for small intestinal (0–6 h), colon (6–24 h) and total tract (0–24 h) permeability, respectively).
Glucose tolerance
Prebiotic treatment had no significant effect on glucose, insulin and C-peptide fasting concentrations or responses during IVGTT compared with placebo (Table 2). The change in glucose effectiveness at zero insulin in the placebo group was significantly different from the prebiotic group.
Table 2 Glucose tolerance outcomes at baseline and after 12 weeks of supplementationFootnote * (Mean values with their standard errors; medians and interquartile ranges (IQR); n 13 for placebo group and n 14 for prebiotic group)

tAUC, total AUC; iAUC, incremental AUC; AIRg: acute insulin response to glucose; DI, disposition index; SI, insulin sensitivity; GEZI, glucose effectiveness at zero insulin; IR, insulin resistance; HOMA, homoeostasis model assessment; %B, % β-cells; %S, % sensitivity.
* There were no differences in baseline (Pre) values between groups (P>0·05, unpaired t test or Mann–Whitney test).
† The P-value is for the comparison of the change between groups with Pre value as covariate (ANCOVA).
‡ n 15 for placebo group.
§ Significant within-group change (P<0·05, paired t test or Wilcoxon’s matched pairs signed-rank test).
|| ANCOVA performed on log-transformed values.
¶ n 14 for placebo group.
Inflammatory markers and lipids
There were no significant effects of prebiotic treatment on inflammatory markers, LPS or lipids, although the prebiotic tended to reduce total cholesterol and LDL-cholesterol (online Supplementary Table S1).
Dietary assessment
At baseline, the energy intake in the prebiotic group was 8929 (sem 538) kJ/d with percentage of energy obtained from carbohydrate, sugar, fat, SFA and protein of 42·1 (sem 2·5), 14·5 (sem 1·7), 36·6 (sem 1·5), 12·5 (sem 0·8) and 15·7 (sem 0·9) %, respectively. In the placebo group, the mean daily energy intake was 8683 (sem 581) kJ and carbohydrate, sugar, fat, SFA and protein provided 40·0 (sem 1·5) %, 14·3 (sem 1·0) %, 37·7 (sem 1·5) %, 12·1 (sem 0·4) % and 16·8 (sem 0·8) % of total energy, respectively. The percentage dietary energy from protein increased by 1·1 % in the placebo group, and this was significantly different from that observed in the prebiotic group (online Supplementary Table S2). No other significant differences in dietary intakes were observed between groups.
Gut microbiota composition
Prebiotic fibre treatment did not induce significant changes in diversity, evenness (the relative abundance of species) and richness (the number of species per sample) indices when compared with placebo. However, bacterial diversity, as assessed by the Shannon and inverse Simpson indices, and richness increased significantly within the prebiotic group (online Supplementary Table S3).
Faecal bacterial DNA extraction was unsuccessful (DNA concentration <50 ng/µl) for some samples, resulting in n 11 in the prebiotic group and n 12 in the placebo group for the qPCR data set. After removing samples with <400 bp, the metagenomics data set consisted of n 7 in the prebiotic group and n 9 in the placebo group.
Consistent with previous reports on composition of the gut microbiota in humans, Bacteroidetes and Firmicutes were the two dominant phyla followed by Proteobacteria, unclassified bacteria and Actinobacteria (data not shown). Bacterial community structure in the treatment groups changed only slightly during the study, but the change was greater in the prebiotic group, as can be observed in the non-metric multidimensional scaling (NMDS) plot (online Supplementary Fig. S1(A)). The change in the placebo group was mainly because of the changes in metformin-treated patients (online Supplementary Fig. S1(B)). However, comparison of bacteria abundances at all taxonomic levels did not reveal any significant effect of treatment when adjusted for multiple testing (data not shown). Nonetheless, permutation ANOVA showed a trend towards an effect of treatment (P=0·099) at the OTU level. When metformin was included as a cofactor, metformin had a significant effect on bacterial community structure at the genus level (R 2=0·084, P=0·009), whereas only a trend was detected when the analysis was performed on OTU (R 2=0·039, P=0·078).
Quantification of bacterial groups by quantitative real-time PCR
Prebiotic treatment had no significant effect on Bifidobacterium or any of the other bacteria measured (online Supplementary Fig. S2). Bifidobacterium levels increased in both groups; however, the change within the prebiotic group was greater and close to significance (P=0·0582).
Correlations between changes in bacteria, clinical outcomes and dietary intakes
As an a priori aim was to investigate the role of prebiotic fibre intake specifically for hypothesis generation, correlations were calculated for each treatment group separately. The correlations differed between the two groups as can be observed from the different patterns in the heat maps (online Supplementary Fig. S3(A–E)). Changes in large bowel permeability (51Cr-EDTA 6–24 h excretion) were positively correlated with bacterial changes at all taxonomic levels in the prebiotic group. The strongest correlations were for Verrucomicrobia, Euryarchaeota and Methanobacteria (online Supplementary Fig. S3(A, B)); Rikenellaceae and unclassified Clostridiales (online Supplementary Fig. S3(D)); and six genera, including Alistipes, Shigella and Flavonifractor (online Supplementary Fig. S3(E)). Furthermore, changes in small intestinal and total IP (51Cr-EDTA 0–6 h and 0–24 h excretion, respectively) correlated positively with changes in Enterobacteriaceae measured by qPCR (r 0·527, P=0·024, adjusted P=0·51 for both small intestinal and total tract permeability) in the prebiotic group. In contrast, only few bacteria correlated with changes in glucose tolerance outcomes; Actinobacteria and Bifidobacterium correlated positively and Veillonellaceae and Clostridium cluster XVIII inversely with glucose total AUC (tAUC) (online Supplementary Fig. S3(A–D)). Unclassified Enterobacteriaceae correlated positively with fasting glucose, insulin sensitivity, high-sensitivity C-reactive protein and waist circumference (online Supplementary Fig. S3(D)).
In the prebiotic group the strongest correlations between bacteria and inflammatory markers were observed for sCD14, which correlated inversely with Verrucomicrobia and unclassified bacteria, Erysipelotrichales and Verrucomicrobiales, Verrucomicrobiacea, Lactobacillaceae and Erysipelotrichaceae (online Supplementary Fig. S3(A, C, D)). Actinobacteria and Firmicutes correlated positively with IL-6 and TNF-α, respectively (online Supplementary Fig. S3(A)). Furthermore, IL-6 correlated positively with Bifidobacterium and negatively with Veillonellaceae and Dialister (online Supplementary Fig. S3(C, D, E)). Changes in small IP correlated with glucose response (incremental AUC) and carbohydrate energy percentage (r −0·429, P=0·033 for both) and colon IP correlated with protein intake (r 0·464, P=0·021) in the prebiotic group. However, because of the small sample size, apart from the association between Veillonellaceae and IL-6 and glucose tAUC (r −0·90, adjusted P=0·042 for both) none of these correlations in the prebiotic group were statistically significant after adjustment for multiple testing.
Discussion
In this study, 12 weeks of prebiotic fibre supplementation did not have a significant beneficial effect on glucose tolerance outcomes in individuals with well-controlled T2D. Although there was a decrease in the IP in the prebiotic group, this was not statistically significant. Because of the number of patients presenting with permeability values within the normal range being higher than expected based on our previous work (50 v. 28 %)( Reference Horton, Wright and Smith 3 ), in future, it would be deemed necessary to test the role of prebiotics in those with a demonstrated impairment in barrier function to assess the true functionality of this dietary fibre.
Bifidobacterium levels increased in both treatment groups, although there was a trend towards post-intervention levels being higher in the prebiotic group. GOS has previously been shown to increase bifidobacteria levels, although it was noted that some volunteers were non-responders( Reference Vulevic, Juric and Walton 23 , Reference Davis, Martinez and Walter 24 , Reference Davis, Martinez and Walter 33 , Reference Walton, van den Heuvel and Kosters 34 ) and one study did not find a significant bifidogenic effect of GOS compared with placebo treatment( Reference Alles, Hartemink and Meyboom 35 ). Interestingly, others have reported a poorer bifidogenic effect of GOS in males and overweight individuals( Reference Walton, van den Heuvel and Kosters 34 ). However, other factors may play a role in these negative findings including the type and dosage of GOS administered, background diet, as well as the methods of analysis of Bifidobacterium ( Reference Davis, Martinez and Walter 33 ). As for the background diet, particularly the relatively high dietary fibre intake (>20 g/d) in this cohort may have diminished the effect of the prebiotic supplement.
We used a dose of 5·5 g prebiotic/d, which may be considered to be low compared with other studies in which doses of 10 g or more prebiotic were consumed( Reference Pourghassem Gargari, Dehghan and Aliasgharzadeh 16 , Reference Alles, de Roos and Bakx 18 , Reference Luo, Van Yperselle and Rizkalla 19 ). A duration of 12 weeks may not have been sufficient to elicit a significant effect on clinical outcomes, although it would have been ample time for changes in the microbiota to become apparent. Resistant starch (which is also a prebiotic) improves first-phase insulin secretion and insulin sensitivity in individuals at risk of T2D within this time scale( Reference Bodinham, Smith and Wright 26 , Reference Johnston, Thomas and Bell 36 ); however, it shows less efficacy in those already with T2D( Reference Bodinham, Smith and Thomas 37 ). An unexpected finding was a decrease in first-phase insulin secretion and an increase in HbA1c in both groups in addition to an increase in fasting glucose within the prebiotic group. This suggests that short-term treatment with a low-dose prebiotic fibre does not prevent further deterioration of key clinical parameters in T2D. The metabolic derangements in established T2D may be difficult to reverse, as shown by the fact that prebiotic supplementation( Reference Alles, de Roos and Bakx 18 , Reference Luo, Van Yperselle and Rizkalla 19 , Reference Bodinham, Smith and Thomas 37 ) does not improve glucose control in T2D, whereas a high efficacy is shown in metabolic syndrome.
Metformin had a significant effect on the intestinal bacterial composition at the genus level, although it only explained a small part (<10 %) of the variation in bacterial composition. Others have recently demonstrated a profound effect of metformin on intestinal bacterial community, bile acids, gut architecture, intestinal glucose utilisation, as well as circulating glucagon-like peptide 1, LBP and LPS( Reference Karlsson, Tremaroli and Nookaew 9 , Reference Napolitano, Miller and Nicholls 38 – Reference Forslund, Hildebrand and Nielsen 43 ). The effect of metformin on glucose control may partly be mediated by these intestinal effects; the increase in the mucin-degrading bacteria Akkermansia muciniphila following metformin treatment is thought to be beneficial( Reference Everard, Belzer and Geurts 15 , Reference Shin, Lee and Lee 40 ). Prebiotics have been shown to increase A. muciniphila in mice( Reference Everard, Belzer and Geurts 15 ); however, we did not observe significant changes in A. muciniphila levels following prebiotic treatment. However, it is a limitation of this study that all thirteen participants for whom bacterial data were available in the prebiotic group were on metformin, whereas only seven participants in the placebo group were on metformin. It seems plausible that metformin may have masked the effects of the prebiotic in the present study, and it is a possible explanation underlying the discrepancy with both animal work and metabolic syndrome, as metformin treatment would not be administered in animal models of T2D.
The fact that the cohort in this study consisted of patients with well-controlled T2D may also play a role. Inflammatory markers were generally low in this group, and this may have been because of a favourable combination of lifestyle factors and medication. However, inflammatory markers are often low in patients with T2D. This may be because some of the antihypertensive and lipid-lowering medications taken by the patients in this study have anti-inflammatory properties and these types of medications may also influence gut bacterial composition( Reference Catry, Pachikian and Salazar 44 ). No clear links between IP and intestinal bacteria were found in this study. The positive correlation between Enterobacteriaceae and 51Cr-EDTA recovery was not significant after adjustment for multiple testing, although it has been useful in hypothesis generation for future work. Others have suggested that a potential link exists between gut health and Enterobacteriaceae because of endotoxin-producing opportunistic pathogens in this bacterial family( Reference Walker and Lawley 45 ). Nevertheless, we found a significant inverse association between changes in Veillonellaceae and IL-6 and glucose tAUC, suggesting a link between this bacterial family, inflammation and glucose response. Veillonellaceae comprises several acetate and propionate producers( Reference Kettle, Louis and Holtrop 46 ), and it has been suggested that SCFA may mediate some of the beneficial effects of prebiotics on host metabolism( Reference Byrne, Chambers and Morrison 47 ). The limitations in this study are primarily related to the small sample size, which makes it difficult to detect subtle effects of a low dose of prebiotic in a heterogeneous study cohort and the potential confounding effects of various medications. In this study, a decision was made at the outset to include numerous clinical and bacterial outcomes, in order to be hypothesis generating for future more focused clinical studies.
In conclusion, supplementation with a low-dose prebiotic for 12 weeks in metformin-treated T2D patients did not improve glucose control; this is now in line with other work showing lack of efficacy of dietary fibres in the treatment of T2D in contrast to their beneficial role in T2D prevention( Reference Bodinham, Smith and Thomas 37 ). However, our study was limited by the small sample size. Before adjustment for multiple testing, many significant associations between changes in intestinal bacteria and clinical outcomes were observed during this study, providing focus and avenues for further work. The commonly used drug metformin is now known to be a significant confounder in the study of bacterial populations in T2D and must be accounted for in future work in this cohort.
Acknowledgements
The authors acknowledge the staff at the CEDAR Centre, Royal Surrey County Hospital, and Dr Caroline Bodinham and Dr Martin Whyte, University of Surrey, for their assistance in the clinical experiments, Amandine Bever, Catholic University of Louvain, for assistance in ELISA and to the study participants for their time and support. The authors also acknowledge Clasado Ltd for providing the supplements.
This study was funded by an EFSD clinical research grant. The research was supported by the National Institute for Health Research Clinical Research Network: Kent, Surrey and Sussex.
M. D. R.: obtained the funding, designed and supervised the research. C. P., E. G., F. H., P. H. and M. D. R.: conducted the clinical experiments. R. J. E.: performed the next-generation sequencing. E. J.: performed the DNA extraction. O. D.: performed qPCR. T. D. and P. D. C.: performed inflammatory marker and LPS measurements. C. P., U. Z. I. and H. W.: analysed the data. J. W.: provided medical supervision. C. P., U. Z. I., R. J. E., O. D. and M. D. R.: wrote the manuscript. R. L. R., G. R. G., O. D. and P. D. C.: edited the manuscript. M. D. R. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
The authors declare that there is no duality of interest associated with this manuscript.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114516004086







