Introduction
In the 20th century the north-east area of São Paulo state, Brazil, changed drastically (Dean, Reference Dean1996; Martinelli & Filoso, Reference Martinelli and Filoso2008). The development of urban centres, expansion of sugar cane and eucalyptus plantations, and extensive enlargement of the road network resulted in considerable habitat loss and fragmentation of natural vegetation cover. Despite this intensive transformation some large carnivore species, such as the puma Puma concolor, still inhabit the area (Miotto et al., Reference Miotto, Rodrigues, Ciocheti and Galetti2007; Lyra-Jorge et al., Reference Lyra-Jorge, Ciocheti and Pivello2008). How these carnivores are able to survive in the remnant habitat fragments, and their population sizes, require study. Estimation of the size of carnivore populations is important for conservation management, especially in human-dominated landscapes, and monitoring this parameter provides managers with a measure of the effectiveness of conservation actions (Caughley, Reference Caughley1994; Karanth et al., Reference Karanth, Nichols, Kumar and Hines2006; Kelly et al., Reference Kelly, Noss, Di Bitetti, Maffei, Arispe and Paviolo2008).
Mark–recapture methods have been used to estimate population size of large carnivores, based on camera-trap photographs (Karanth & Nichols, Reference Karanth and Nichols1998; Trolle & Kéry, Reference Trolle and Kéry2003; Silver et al., Reference Silver, Ostro, Marsh, Maffei, Noss and Kelly2004; Trolle et al., Reference Trolle, Noss, De Lima and Dalponte2007; Kelly et al., Reference Kelly, Noss, Di Bitetti, Maffei, Arispe and Paviolo2008; Paviolo et al., Reference Paviolo, DiBlanco, De Angelo and DiBitetti2009; Silveira et al., Reference Silveira, Jácomo, Astete, Sollmann, Torres, Furtado and Marinho-Filho2009) or radio-telemetry tracking data (Franklin et al., Reference Franklin, Johnson, Sarno and Iriarte1999; Logan & Sweanor, Reference Logan and Sweanor2001; Soisalo & Cavalcanti, Reference Soisalo and Cavalcanti2006). As an alternative, noninvasive genetic sampling has become a powerful tool for studying and monitoring elusive and low-density species (Waits & Paetkau, Reference Waits and Paetkau2005; Schwartz et al., Reference Schwartz, Luikart and Waples2007; Janečka et al., Reference Janečka, Jackson, Yuquang, Diqiang, Munkhtsog, Buckley and Murphy2008). DNA from sources such as hair or faeces can be used as molecular tags in mark–recapture population censuses (Bellemain et al., Reference Bellemain, Swenson, Tallmon, Brunberg and Taberlet2005; Prugh et al., Reference Prugh, Ritland, Arthur and Krebs2005; Boulanger et al., Reference Boulanger, Kendall, Stetz, Roon, Waits and Paetkau2008; De Barba et al., Reference De Barba, Waits, Genovesi, Randi, Chirichella and Cetto2010). In this context collecting noninvasive samples is equivalent to capturing the animal that deposited the genetic sample. The probability of encountering faeces can therefore be referred to the capture probability. Because each animal has a unique multilocus genotype, each individual can be identified. Closed or open-population models can then be applied, depending on the length of the sampling period, and births, deaths, emigration and immigration can be used to estimate population parameters such as abundance, survival and recruitment.
The puma is the most widely distributed large terrestrial mammal in the Americas (Iriarte et al., Reference Iriarte, Franklin, Johnson and Redford1990). It has been studied extensively in North America (Ross & Jalkotzy, Reference Ross and Jalkotzy1992; Roelke et al., Reference Roelke, Martenson and O'Brien1993; Lindzey et al., Reference Lindzey, Van Sickle, Ackerman, Barnhurst, Hemker and Laing1994; Beier et al., Reference Beier, Choate and Barret1995; Ernest et al., Reference Ernest, Penedo, May, Syvanen and Boyce2000; Logan & Sweanor, Reference Logan and Sweanor2001; Anderson et al., Reference Anderson, Lindzey and McDonald2004; McRae et al., Reference McRae, Beier, Dewald, Huynh and Keim2005) but not in such detail in South America. Puma population sizes and densities have been estimated in a few studies (Franklin et al., Reference Franklin, Johnson, Sarno and Iriarte1999, in Patagonia, Chile; Kelly et al., Reference Kelly, Noss, Di Bitetti, Maffei, Arispe and Paviolo2008, in Bolívia, Argentina and Belize; Paviolo et al., Reference Paviolo, DiBlanco, De Angelo and DiBitetti2009, in the Green Corridor of Atlantic Forest in Misiones Province, Argentina) but there have been few studies in Brazil (Mazzolli, Reference Mazzolli2010; Negrões et al., Reference Negrões, Sarmento, Cruz, Eira, Revilla and Fonseca2010), especially in human-disturbed areas. Puma research in Brazil has concentrated on food habits or habitat use (Emmons, Reference Emmons1987; Ciocheti, Reference Ciocheti2007; Lyra-Jorge et al., Reference Lyra-Jorge, Ciocheti and Pivello2008), movement (Schaller & Crawshaw, Reference Schaller and Crawshaw1980) and human–puma conflicts (Mazzolli et al., Reference Mazzolli, Graipel and Dunstone2002; Conforti & Azevedo, Reference Conforti and Azevedo2003; Azevedo, Reference Azevedo2008).
Here, we present an estimate of the population size of pumas inhabiting a human-disturbed landscape in the north-east of São Paulo state, based on a DNA mark–recapture method. Using microsatellites we individualized marked and recaptured samples of puma faeces and estimated population abundance by applying an open population model framework.
Study area
The study area encompasses c. 260 km2 and is located in the Luís Antônio and Santa Rita do Passa Quatro municipalities, in north-east São Paulo state, Brazil (Fig. 1). The area is characterized as transitional between Cerrado and semi-deciduous Atlantic forest. There are two protected areas 3 km apart, the Jataí Ecological Station and the Vassununga State Park, in addition to some habitat patches on private properties. The 9,000 ha Jataí Ecological Station is the largest protected area in the state with continuous Cerrado vegetation. The 2,070 ha Vassununga State Park contains six distinct patches of Cerrado and semi-deciduous forest (Korman, Reference Korman2003). In our sampling in Vassununga State Park we concentrated on the largest patch (Pé de Gigante, 1,210 ha), as the smaller patches are surrounded by intensive human activity and unlikely to be used by pumas. Both protected areas are surrounded by sugar cane crops, cattle ranches, eucalyptus plantations, dirt roads and highways but still retain high faunal diversity (Talamoni et al., Reference Talamoni, Motta, Dias, Santos and Pires2000).

Fig. 1 The location of Jataí Ecological Station and the Pé de Gigante forest patch (the latter a part of Vassununga State Park) in São Paulo state, showing the roads where the surveys were made and the locations where faeces were collected. The insets show the location of the main map in Brazil (left) and in north-east São Paulo state (right).
Methods
Sample collection
From 2004 to 2008 we conducted a noninvasive puma monitoring project in the study area (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012), collecting faeces preferentially in the dry seasons (May–August) of each year, with the exception of 2008, when we conducted monthly 2-day field excursions from March to October. For the analysis described here we used only the 2008 data because they were collected at regular intervals and on the same roads. There were eight sampling occasions, and each 2-day excursion was considered one sampling occasion. In the study area fragments and plantations are either surrounded or intersected by low-traffic dirt roads. Our sampling scheme encompassed these roads, covering c. 200 km in each field trip (Fig. 1). Pumas tend to use dirt roads to move from one place to another (Dickson et al., Reference Dickson, Jenness and Beier2005) and, especially in the study area, they commonly defecate on these roads (authors, pers. obs.). Samples were collected exclusively from these roads. All fieldwork was conducted by car and by two researchers. Based on tracks and the diameter and morphology of faeces we collected 37 potential puma faecal samples, and we recorded the locations of all collections, using a global positioning system. Samples were stored in sterile, preservative-free plastic tubes without any conservation solution and kept at −22 °C in the laboratory until the DNA extraction was performed.
DNA extraction
Faecal DNA was extracted using the QIAmp DNA Stool Mini Kit (Qiagen) or PSP Spin Stool DNA Kit (Invitek), following manufacturer's recommendations. For the blood and tissue DNA extractions used as reference sequences in the genetic analysis we followed the phenol/chloroform/isoamylic alcohol protocol described in Sambrook et al. (Reference Sambrook, Fritsch and Maniatis1989). We collected these samples directly from pumas killed or injured in collisions with vehicles in the region and from the government agency Centro Nacional de Pesquisa e Conservação de Mamíferos Carnívoros, which has a blood/tissue bank.
Species identification
To confirm the species of the collected faecal samples we amplified and sequenced a 146 bp portion from the cytochrome b gene of the mitochondrial DNA (mtDNA), using primers described by Farrell et al. (Reference Farrell, Roman and Sunquist2000). Details of the amplification, sequencing and analysis procedures are described in Miotto et al. (Reference Miotto, Rodrigues, Ciocheti and Galetti2007).
Individual identification
To individualize each faecal sample we amplified a set of seven species-specific microsatellite loci, with primers developed by Kurushima et al. (Reference Kurushima, Collins and Ernest2006): Pco C108 (a tetranucleotide-repeat microsatellite locus), and Pco B010, Pco B210, Pco A339, Pco A208, Pco A216 and Pco B003 (dinucleotide-repeat microsatellite loci). Primers were marked with universal fluorescent M13 tails following Schuelke (Reference Schuelke2000). Each PCR reaction (15 μL) contained 7.5 μL of GoTaq Master Mix (Promega), containing 1×buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 1 unit Taq polymerase, 8 pmol of reverse primer, 2 pmol of forward primer, 8 pmol of M13 sequence marked with the 6-FAM fluorophore, and 150 μg ml−1 of BSA. The remaining volume of the 15 μL reaction comprised DNA. Amplifications were performed in a PTC-100 Thermocycler (MJ Research) according to the following protocol (for all primer pairs): an initial denaturation cycle at 94 °C for 5 minutes, 40 cycles at 92 °C for 1 minute, 48 °C for 1 minute, and 72 °C for 1 minute, and a final 30-minute extension at 72 °C. Negative controls were included in all reactions to monitor possible contamination. The resulting genotypes were analysed in a MegaBACE ET-550R Size Standard automatic sequencer (GE Healthcare) with Genetic Profiler (GE Healthcare). We conducted individual identification of genotypes using Gimlet (Valière, Reference Valière2002). To prevent misidentification as a result of allelic dropout, in which one of the two alleles in a heterozygous individual is not detected (Taberlet et al., Reference Taberlet, Griffin, Goossens, Questiau, Manceau and Escaravage1996), we genotyped homozygote samples in 3–5 independent PCR reactions and genotyped heterozygotes twice. For individual identification only samples that were successfully genotyped for at least five loci were included in the analysis. To quantify the power to discriminate individuals with the microsatellite loci used we determined the identity probability (P (ID)); i.e. the probability of two individuals in a population randomly sharing identical genotypes for all the analysed loci (Paetkau et al., Reference Paetkau, Waits, Clarkson, Craighead, Vyse, Ward and Strobeck1998; Waits et al., Reference Waits, Luikart and Taberlet2001). The P (ID) values were calculated based on the allele frequencies of unique genotypes for each locus, in Gimlet (Valière, Reference Valière2002), and then multiplied by the total number of loci to obtain a total P (ID) (Paetkau et al., Reference Paetkau, Waits, Clarkson, Craighead, Vyse, Ward and Strobeck1998). Because we expected to sample related animals in the study area (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012) we calculated both the P (ID)unbiased and P (ID)sib equations, taking into account the population size and the presence of related individuals, respectively (Waits et al., Reference Waits, Luikart and Taberlet2001). We determined the total and per-locus genotyping error rates (allelic dropout) by dividing the number of detected errors by the number of cases in which an error might have been detected (i.e. the total number of genotyping reactions). Finally, we investigated null allele and stutters occurrence with Micro-Checker 2.2.3 (Van Oosterhout et al., Reference Van Oosterhout, Hutchinson, Wills and Shipley2004)
Sex determination
To identify the sex of the individuals sampled we amplified a portion of the amelogenin gene present in both sex chromosomes, using primers described by Pilgrim et al. (Reference Pilgrim, McKelvey, Riddle and Schwartz2005). In this gene fragment males have a 20 bp deletion in the Y-chromosome copy and consequently produce PCR products of different sizes, whereas females amplify fragments of the same size. To prevent false positive for females we repeated each reaction three times.
Abundance of pumas
After individualizing puma faecal samples, a capture–recapture history of each animal was established for the 8-month sampling period. To estimate abundance (
![]() $\hat N$
), we opted for an open population model as we assumed that animals could leave or immigrate into the study area during the sampling period. Pumas are vagile and have large home ranges and we had previous information that suggested some may frequently disperse throughout the matrix (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012). We estimated
$\hat N$
), we opted for an open population model as we assumed that animals could leave or immigrate into the study area during the sampling period. Pumas are vagile and have large home ranges and we had previous information that suggested some may frequently disperse throughout the matrix (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012). We estimated
![]() $\hat N$
with the Jolly–Seber probability model (Jolly, Reference Jolly1965; Seber, Reference Seber1965), using the POPAN formulation (Schwarz & Arnason, Reference Schwarz and Arnason1996), implemented in MARK (White & Burnham, Reference White and Burnham1999). This open population model assumes the existence of a ‘super-population’ (N) in which unmarked animals have the same probability of capture as marked animals in the population (assumption of equal catchability), survival rates are homogeneous for marked and unmarked animals, the sampled area is constant, and there is no loss of mark (genetic tag in our case) during the sampling period. The parameters of this model are p
i, the probability of capture of marked and unmarked individuals on occasion i, ф
i
, the probability of survival of marked and unmarked individuals from occasion i to i + 1, and b
i
, the probability of a new animal entering the population between occasions i and i + 1. To test the validity of the above assumptions a goodness of fit test was performed using RELEASE v. 3.0 (Burnham et al., Reference Burnham, Anderson, White, Brownie and Pollock1987) and by accessing the deviance of 1,000 bootstrapped simulations based on the global time-dependent Cormack–Jolly–Seber model (Dunstan et al., Reference Dunstan, Bradshaw and Marshall2011). We selected between eight models, all nested inside the global model, allowing for temporal variation in all three parameters [p(t)ф(t)b(t)N(.)]. The tested models are described in Table 1. To guarantee numerical convergence a multinomial link function was used for the bi parameters (constraining a set of real parameters to sum to 1.00), and in the case of the abundance parameter an identity link function was used, permitting the parameter to have values outside the [0, 1] interval. Models were ranked using the Akaike information criterion (AICc) adjusted for small population sizes. The model with the lowest AICc value was considered the model that best balanced bias and precision (White & Burnham, Reference White and Burnham1999) and, therefore, the most appropriate model for accurate estimation of abundance.
$\hat N$
with the Jolly–Seber probability model (Jolly, Reference Jolly1965; Seber, Reference Seber1965), using the POPAN formulation (Schwarz & Arnason, Reference Schwarz and Arnason1996), implemented in MARK (White & Burnham, Reference White and Burnham1999). This open population model assumes the existence of a ‘super-population’ (N) in which unmarked animals have the same probability of capture as marked animals in the population (assumption of equal catchability), survival rates are homogeneous for marked and unmarked animals, the sampled area is constant, and there is no loss of mark (genetic tag in our case) during the sampling period. The parameters of this model are p
i, the probability of capture of marked and unmarked individuals on occasion i, ф
i
, the probability of survival of marked and unmarked individuals from occasion i to i + 1, and b
i
, the probability of a new animal entering the population between occasions i and i + 1. To test the validity of the above assumptions a goodness of fit test was performed using RELEASE v. 3.0 (Burnham et al., Reference Burnham, Anderson, White, Brownie and Pollock1987) and by accessing the deviance of 1,000 bootstrapped simulations based on the global time-dependent Cormack–Jolly–Seber model (Dunstan et al., Reference Dunstan, Bradshaw and Marshall2011). We selected between eight models, all nested inside the global model, allowing for temporal variation in all three parameters [p(t)ф(t)b(t)N(.)]. The tested models are described in Table 1. To guarantee numerical convergence a multinomial link function was used for the bi parameters (constraining a set of real parameters to sum to 1.00), and in the case of the abundance parameter an identity link function was used, permitting the parameter to have values outside the [0, 1] interval. Models were ranked using the Akaike information criterion (AICc) adjusted for small population sizes. The model with the lowest AICc value was considered the model that best balanced bias and precision (White & Burnham, Reference White and Burnham1999) and, therefore, the most appropriate model for accurate estimation of abundance.
Table 1 Model selection results for the POPAN parameterized models for puma Puma concolor capture data in north-east São Paulo state (Fig. 1).

* ф, survival probability; p, capture probability; b, probability of an individual leaving or entering the study area during the sampling period; (.), constancy; (t), temporal variation
Results
Of the 37 faecal samples collected mtDNA was amplified in 27 samples (84.37%). By comparing the sequenced mtDNA fragments we identified 24 puma faeces. Three samples belonging to ocelots Leopardus pardalis, a sympatric species present in the area, were discarded from further analysis. Genotyping success for the set of seven microsatellite loci was > 80%. We quantified a general rate of 6.56% for allelic dropout and estimated a total P (ID)unbiased of 6 × 10−9 and a total P (ID)sib of 0.001. Our data presented no evidence of null alleles or stutter peaks. The range size, number of alleles per locus, and PID values per locus are described in Table 2. From the 24 samples we identified 15 different pumas. Table 3 presents their mark–recapture data (encounter history).
Table 2 The range size, number of genotyped samples, number of alleles, probability of identity (unbiased and with sibs), and per locus and total allelic dropout rate for the seven microsatellite loci analysed.

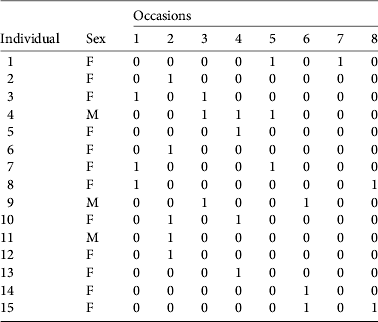
Table 3 Encounter history (1, detected; 0, not detected) of 15 individualized female (F) and male (M) pumas in the study area, from eight periods of faecal collection.
No evidence of overdispersion was found for the global model and therefore our models were considered to adequately fit the data and no important assumptions were being violated. Model selection in MARK resulted in {Φ(.)p(.)b(t)N(.)} (constant capture probability, constant survival rate, and temporal variation in the probability of new animals entering the population between sampling occasions) being the most appropriate model (AICc = 112.399; Table 1). The high ΔAICc values for the remaining models allowed us to distinguish the first model as the only one that sufficiently supported the data and, therefore, this was the only model appropriate for obtaining further estimates. Estimated puma abundance using this model was 23.81 ± SE 6.22 (95% CI 17.53–45.66). The capture probabilities for this model were constant and equal to 0.19 ± SE 0.06 (95% CI 0.098–0.339).
Discussion
Noninvasive genetic analysis
The estimated values of probability of identity were low for the seven loci analysed (P (ID)unbiased of 6 × 10−9 and P (ID)sib of 0.001). Both values are considered satisfactory for individual identification in noninvasive genotyping (Waits et al., Reference Waits, Luikart and Taberlet2001) and indicate that the analysed loci successfully distinguished each individual, even in the presence of closely related animals (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012). Because of low DNA quality and quantity, microsatellite analyses of noninvasive samples, such as faeces, are commonly affected by genotyping errors, such as allelic dropout or false allele amplification, and consequently the identification of individuals and the estimates of population size could be biased (Taberlet et al., Reference Taberlet, Griffin, Goossens, Questiau, Manceau and Escaravage1996, Reference Taberlet, Waits and Luikart1999; Waits et al., Reference Waits, Luikart and Taberlet2001; Broquet & Petit, Reference Broquet and Petit2004; Prugh et al., Reference Prugh, Ritland, Arthur and Krebs2005). To avoid errors we established conditions to obtain consistent genotypes: we genotyped heterozygotes twice and confirmed the genotypes of homozygotes with 3–5 independent PCR reactions, we included a locus in our analysis only if > 80% of samples were successfully genotyped, and in the final analysis we only included those samples that were genotyped for at least 80% of all the analysed loci.
Unlike other felid species pumas have no pelage patterns, such as the rosettes that are features of jaguars and ocelots (Trolle & Kéry, Reference Trolle and Kéry2003; Silver et al., Reference Silver, Ostro, Marsh, Maffei, Noss and Kelly2004; Trolle et al., Reference Trolle, Noss, De Lima and Dalponte2007; Silveira et al., Reference Silveira, Jácomo, Astete, Sollmann, Torres, Furtado and Marinho-Filho2009) or the stripes of tigers (Karanth & Nichols, Reference Karanth and Nichols1998; Karanth et al., Reference Karanth, Nichols, Kumar and Hines2006) that could allow reliable individual identification in camera-trap studies. Kelly et al., (Reference Kelly, Noss, Di Bitetti, Maffei, Arispe and Paviolo2008) proposed a protocol to identify individual pumas based on body patterns analysed by distinct researchers. This protocol was later used by Paviolo et al. (Reference Paviolo, DiBlanco, De Angelo and DiBitetti2009) but some subjectivity in individual identification remained once each researcher identified a distinct number of animals (Kelly et al., Reference Kelly, Noss, Di Bitetti, Maffei, Arispe and Paviolo2008). Mazzolli (Reference Mazzolli2010) and Negrões et al., (Reference Negrões, Sarmento, Cruz, Eira, Revilla and Fonseca2010) also based their estimates of puma population size on photographs but both studies were subject to some degree of subjectivity. Foster & Harmsen (Reference Foster and Harmsen2011) listed a number of biases that can compromise the individual identification of species that lack individually identifiable natural markers. Thus, despite some inherent challenges in the technique, mainly in tropical humid and hot regions (Norris & Michalski, Reference Norris and Michalski2010), as long as a rigid amplification protocol against genotype misidentification is applied (i.e. reducing allelic dropout or false allele occurrence), noninvasive genetic analysis may guarantee more accuracy in individual identification, especially for species without evident pelage patterns, and thus avoid potential under- or overestimation of population size.
Population size estimators
The best model in our selection analysis supports temporal variation in recruitment (i.e. variation in the pattern by which new animals enter the population, either by births or immigration) and implies constancy of survival and capture probability. The support for this model was substantial, as the ΔAICc of the second best model was > 2 (Burnham & Anderson, Reference Burnham and Anderson1998). The model is consistent with the species’ biology as pumas do not have a specific reproductive season, with mating occurring throughout the year (Logan & Sweanor, Reference Logan and Sweanor2001). Male pumas tend to disperse from natal areas whereas females may exhibit phylopatry or disperse over short distances (Logan & Sweanor, Reference Logan and Sweanor2001). Therefore, the detected variation could be a direct response to reproduction, with the entry of males into the area for mating and the subsequent entry of newborn males and females, or a response of subadults leaving the area after the maternal care period, which occurs at 16–22 months of age (Logan & Sweanor, Reference Logan and Sweanor2001). Unfortunately, by using molecular markers we were not able to establish age structure and, consequently, could not distinguish cubs, juveniles or adults. In a previous study (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012), however, we detected few males and many resident females, suggesting that males may be more responsible for the flow of individuals into and out of the area.
Our abundance estimate represents the entire population (males and females), as grouping the data would have weakened the analysis through over-parameterization of models. The models used for estimating abundance showed no evidence of lack of fit to the data, which reinforced the accuracy of the abundance estimates. Our sample size was small but, in addition to being constant, P (0.19 ± SE 0.06) was higher than the mean reported in other felid studies (0.13 ± SE 0.09; Foster & Harmsen, Reference Foster and Harmsen2011).
To compare our results with other studies in South America (Franklin et al., Reference Franklin, Johnson, Sarno and Iriarte1999; Kelly et al., Reference Kelly, Noss, Di Bitetti, Maffei, Arispe and Paviolo2008; Paviolo et al., Reference Paviolo, DiBlanco, De Angelo and DiBitetti2009), including Brazil (Mazzolli, Reference Mazzolli2010; Negrões et al., Reference Negrões, Sarmento, Cruz, Eira, Revilla and Fonseca2010), we would need to estimate density. However, as the population was demographically open we are unable to make such a comparison. For geographically open populations the effective sampled area (Â) is poorly defined relative to the sampled population (White et al., Reference White, Anderson, Burham and Otis1982) and studies using telemetry data have shown that density is overestimated by
![]() $\hat N$
/Â (Soisalo & Cavalcanti, Reference Soisalo and Cavalcanti2006; Dillon & Kelly, Reference Dillon and Kelly2008).
$\hat N$
/Â (Soisalo & Cavalcanti, Reference Soisalo and Cavalcanti2006; Dillon & Kelly, Reference Dillon and Kelly2008).
Puma population size
Estimates of population size and dynamics of the puma are crucial for long-term conservation of the species, especially in human-disturbed landscapes. Our estimates of population size (23.81 ± SE 6.22) is high considering the small size of the study area. There appears to have been a recent increase in the number of pumas in north-east São Paulo state and other regions of Brazil. For example, from 2004 to 2012 in a 1,700 km2 area that encompasses our study area, 26 pumas were killed on roads and 11 were rescued in urban areas as a result of livestock depredation (R.A. Miotto, pers. obs. & unpubl. data). Little is known about the reasons for this possible expansion, or if it will continue, but we believe it is influenced by the characteristics of the area.
In the previous 50 years the area planted with sugar cane in Brazil increased from c. 1.4 to 7 million ha (Martinelli & Filoso, Reference Martinelli and Filoso2008). More than 50% of the country's sugar cane land cover is in São Paulo state, with plantations increasing at an annual rate of c. 85,000 ha (Martinelli & Filoso, Reference Martinelli and Filoso2008). In addition to eucalyptus plantations, pastures and the development of urban centres, this intensive human activity has severely transformed the original vegetation cover of the north-east of the state. The area is now characterized by many small and a few large patches of vegetation surrounded by human-disturbed areas. Despite this transformation pumas are still present in the area, probably as a consequence of their generalist habits.
In general, carnivore densities are positively correlated with prey biomass (Franklin et al., Reference Franklin, Johnson, Sarno and Iriarte1999; Logan & Sweanor, Reference Logan and Sweanor2001; Carbone & Gittleman, Reference Carbone and Gittleman2002; Karanth et al., Reference Karanth, Nichols, Kumar and Hines2006). The two protected areas in our study area are amongst the largest natural refuges for many species of the ungulate prey of pumas (Novack et al., Reference Novack, Main, Sunquist and Labisky2005; Ciocheti, Reference Ciocheti2007) in the north-east of São Paulo state. These protected areas are therefore high-quality habitat patches in the matrix and may allow pumas to concentrate in the area and tolerate overlapping territories (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012). Intraspecific conflict may contribute to puma mortality but when prey availability is high female pumas tend to have smaller home ranges (Logan & Sweanor, Reference Logan and Sweanor2001). Even embedded in a relatively permeable matrix, with eucalyptus plantations and sugar cane crops, pumas may be confined to these protected areas when there are no surrounding patches of similar quality. Fragments surrounding the protected areas are more susceptible to hunting pressure, which leads to decreased prey biomass and abundance (Cullen et al., Reference Cullen, Bodmer and Valladares-Pádua2000, Reference Cullen, Bodmer and Valladares-Pádua2001). In addition, the absence of competitors such as the jaguar Panthera onca may facilitate a greater number of pumas in the study area (coexistence between different felid species may regulate population sizes; Donadio & Buskirk, Reference Donadio and Buskirk2006). The study area may act as a source area of pumas (Miotto et al., Reference Miotto, Cervini, Begotti and Galetti2012); i.e. an area where pumas are resident and raise their cubs, which then disperse as juveniles. Because molecular markers do not allow us to define age structure, we could not recognize cubs in our data set, but their probable presence in the area may have increased the population estimate.
The closest protected area >1,000 ha is c. 70 km from Jataí Ecological Station and Vassununga State Park. To maintain the puma population in this region the priority should be to improve connectivity by establishing new protected areas and increasing the size of existing protected areas such as Jataí Ecological Station and Vassununga State Park. The improvement of habitat quality on private properties, with recovery and conservation of remnant forest fragments in areas occupied by the sugar cane and wood pulp industries, would also improve landscape management for the puma.
Acknowledgements
We thank Fundação de Amparo à Pesquisa do Estado de São Paulo and Conselho Nacional de Desenvolvimento Científico e Tecnológico for their financial support, the Capes agency for a PhD scholarship to RAM and a post-doctoral fellowship to MK (PNPD), the Neotropical Grassland Conservancy for supplying laboratory reagents, Marina Gomes de Figueiredo and Giordano Ciocheti for their help with fieldwork, and two anonymous reviewers for their comments and suggestions.
Biographical sketches
Renata A. Miotto is investigating the genetics and ecology of carnivorous mammals living in fragmented and disturbed areas in Brazil. Marcelo Cervini is a geneticist working on the conservation and evolution of a range of groups, from mammals to bees. Maja Kajin works on population ecology and genetics, ecological modelling, and demography, focusing mainly on Atlantic forest mammals. Rodrigo A. Begotti is studying the conservation of forest resources, and works with landscape ecology and the conservation of medium and large-bodied mammals in fragmented landscapes. Pedro M. Galetti Jr's research group studies the conservation genetics of a diverse range of organisms.






