INTRODUCTION
Diarrhoeal diseases in children aged <5 years are a significant cause of morbidity and mortality in the developing world [1], and in Malawi, they are thought to be the sixth leading cause of mortality which currently stands at 182/1000 live births [2]. Cryptosporidiosis is an acute, self-limiting diarrhoeal illness in immunocompetent patients, but in the immunocompromised, especially those with HIV/AIDS, and the malnourished, the disease can be severe, protracted, debilitating, and may lead to death. In Africa, cryptosporidiosis has been associated with 3·8–26% of all diarrhoeal illness in the paediatric population [Reference Mwachari3–Reference Amadi5], and has been diagnosed in at least 47% of African countries. In Malawi, up to 10% of diarrhoea in children aged <5 years is attributable to Cryptosporidium spp. infection with up to 21% reported in the immunocompromised population [Reference Pavone6; M. Perez, unpublished observations]. Of the 14 presently accepted ‘valid’ species of Cryptosporidium, seven have been found to infect humans to date [Reference Xiao7, Reference Ryan8]. In sub-Saharan Africa alone, four of these species have been identified as causing infection to date, namely C. hominis, C. parvum, C. meleagridis and C. muris [Reference Morgan9–Reference Tumwine13].
In 2003, it was estimated that at least 14% of the population in Malawi were infected with HIV, and that 67% of preschool children in Malawi are chronically malnourished causing many children to fail to reach their full growth and cognitive potentials in later life [14]. In the absence of inexpensive, well tested chemotherapy for cryptosporidiosis, prevention of transmission is the only effective means of reducing the incidence of disease and its subsequent impact on morbidity and mortality [Reference Smith and Corcoran15].
To reduce exposure to infectious Cryptosporidium oocysts in the young and immuno-compromised, both risk factors and the epidemiology of disease must be understood. As all species do not appear to cause infection in humans, and the majority of human infective species cannot be differentiated microscopically because of overlap in their oocyst dimensions, both microscopic and molecular methods, which can discriminate between all valid Cryptosporidium spp., are required to identify potential sources of infection.
Here we present data, accrued over 2 years, of the incidence of Cryptosporidium spp. in children aged <5 years in Malawi, using conventional microscopic and molecular species typing tools. This was part of a larger study which investigated the risk factors associated with childhood cryptosporidiosis and the environmental sources of Cryptosporidium in Malawi between urban and rural areas.
MATERIALS AND METHODS
Sample procurement
We studied two districts of the southern region of Malawi representing both urban (Blantyre) and rural (Chikwawa) settings. Permission to conduct the study was obtained from the National Health Sciences Research Committee (Ministry of Health and Population, Malawi Government). The study was conducted from the main Government hospitals in both areas, and local health clinics.
Stool samples were collected from children aged <5 years with diarrhoea. Diarrhoea was defined as an increase in the frequency and/or change in the consistency (to loose or watery) of the stool as determined by the child's mother/guardian. Samples were procured by medical staff who requested that guardians supply a stool sample from the child in a 30 ml plastic container (Enterprise Containers, Blantyre, Malawi). Guardians who provided baseline information; received advice on reducing and treating diarrhoea and were provided with soap for hand washing, oral rehydration salts for (ORS) treatment. Stool samples were transported in a cool box and stored in the laboratory at 4°C.
Detection methods
Microscopy
Cryptosporidium oocysts were detected in direct faecal smears by the modified Ziehl–Neelsen (mZN) and auramine phenol (AP) methods [Reference Casemore, Armstrong and Sands16]. Subsamples of all stools were stored without preservative at 4°C and subjected to further microscopic and molecular analyses in the United Kingdom (Scottish Parasite Diagnostic Laboratory). Putative mZN and/or AP positive samples were confirmed by immunofluorescence (IF) with a commercially available fluorescein isothiocyanate-labelled monoclonal antibody reactive with exposed epitopes on Cryptosporidium spp. oocysts (FITC-C-mAbs) (Cryptoglo, Waterborne Inc. New Orleans, USA; CryptoCel, TCS Water Sciences, Australia), and the nuclear fluorogen 4′6-diamidino-2-phenyl indole (DAPI) [Reference Grimason17]. Positive samples, viewed at ×400, were scored from negative to +3 [Reference McLaughlin18]. Nomarski differential interference contrast (DIC) microscopy was used to determine internal morphology and fluorescence at ×1000 magnification using an Olympus BH-2 fluorescence microscope equipped with DIC optics. A blue filter (480 nm excitation, 520 nm emission) was used to detect FITC-C-mAb-labelled oocysts and DAPI was detected using a UV filter block (350 nm excitation, 450 nm emission).
Molecular typing
Oocyst concentration and DNA extraction
Oocyst positive and putative positive faecal samples were purified by a modified water–ether concentration method [Reference Bukhari and Smith19, Reference Nichols, Moore and Smith20]. With semi-solid samples, a pea-sized faecal sample was diluted in 200 μl distilled water and emulsified in a 1·5 ml microcentrifuge tube, and for liquid faeces, 200 μl were transferred to a 1·5 ml microcentrifuge tube. Samples were concentrated as previously described [Reference Bukhari and Smith19, Reference Nichols, Moore and Smith20]. Finally the supernatant was aspirated down to 100 μl, the remaining sample resuspended and 5 μl transferred to a welled slide for subsequent FITC and DAPI staining as described above. The remainder of the sample was washed twice, as described above, with lysis buffer [LB; 50 mm Tris–HCl (pH 8), 1 mm EDTA (pH 8), 0·5% SDS].
Where insufficient faecal sample was available or where PCR amplification was inhibited, oocysts were recovered from IF slides which had been prepared at the time of sample procurement. Briefly, coverslips were carefully removed from slides to expose the stained sample. Then, 20 μl LB was added and the dried sample mixed with LB. The resuspended sample was transferred into a microcentrifuge tube and sample mixing was repeated five times to ensure maximum oocyst recovery from the sample well. Where necessary to maximize oocyst recovery, sample volume was increased to 100 μl with LB [Reference Nichols, Campbell and Smith21].
DNA was released from oocysts by freeze-thawing and proteinase K digestion [Reference Nichols and Smith22]. The supernatant (70 μl), containing released DNA, was transferred to a clean (DNAse-/RNAse-free) tube and either used immediately for PCR amplification or stored at –20°C until used.
PCR protocols
Cryptosporidium spp. identity was determined for all samples using the following PCR–RFLP assays:
(a) direct PCR amplification of the 18S rRNA gene locus [Reference Johnson23], co-amplification with an internal control [Reference Nichols24] followed by simultaneous digestion with DraI and AseI (D18S rRNA) [Reference Nichols, Campbell and Smith21];
(b) nested PCR amplification of the 18S rRNA gene locus [Reference Nichols, Campbell and Smith21, Reference Johnson23] followed by simultaneous digestion with DraI and AseI (N18S rRNA);
(c) single tube nested PCR amplification of the Cryptosporidium oocyst wall protein (STN-COWP) gene locus, followed by digestion with TaqI [Reference Homan25].
In addition, when samples failed to amplify, or gave insufficient amplicon for digestion using PCR (c), samples were also subjected to two-step nested (2SN) PCR at the Homan et al. [Reference Homan25] COWP gene locus (N-COWP) or the nested 18S rRNA PCR [Reference Xiao26].
All PCR reactions were performed in a GeneAmp® Model 9700 thermocycler (Applied Biosystems, Foster City, CA, USA) using thin-walled 0·5 ml DNAse-/RNAse-free PCR tubes. PCRs (a) and (b) (18S rRNA gene locus) generate an amplicon of 435 bp. Endonuclease restriction with AseI and DraI produces the following fragments in order by size as they are separated on the gel: C. hominis: 222, 104, 112 bp; C. parvum: 219, 104, 112 bp; C. meleagridis: 171, 104, 112, 47 bp; C. felis: 189, 112, 104, 50 bp and C. muris: 320, 112 bp. PCR (c) generates an amplicon of 640 bp. Endonuclease restriction with TaqI produces the following fragments: C. hominis: 470, 170 bp; C. parvum: 374, 266 bp.
RESULTS
Eight hundred and forty-eight diarrhoeal stool samples (urban n=425, rural n=423) were collected between February 2001 and December 2002, of which 50 samples (rural n=25, urban n=25) were confirmed Cryptosporidium positive by a combination of microscopy and molecular techniques. The efficiency of detection of the various techniques used in this study is shown in Table 1.
Table 1. Results of microscopy and PCR-positive tests for presence of Cryptosporidium sp.

mZN, Modified Ziehl–Neelsen; AP, auramine phenol; IF, immunofluorescence; PCR, polymerase chain reaction.
* χ2=10, d.f.=1, P<0·005.
The species of Cryptosporidium infecting these children are shown in Table 2. C. hominis accounted for 48% of positive samples and C. parvum for 18%. Four samples generated amplicons typical in size and RFLP profiles to that of C. hominis/C. parvum with N18S rRNA. However, further PCR targeting the COWP gene loci and the N18S rRNA locus of Xiao et al. [Reference Xiao26] failed to generate positive PCRs for these four samples. A further sample generated an amplicon typical in size to that of C. andersoni/C. muris and a typical C. andersoni RFLP profile with DdeI, but insufficient sample was available for confirmation. Seven samples did not generate a PCR product despite containing oocysts. Two of these samples were shown to be inhibitory with the use of the internal control. However, no inhibition was evident in the remaining five samples, which were all positive when direct smears were examined by at least two microscopic methods. Examination of the water–ether concentrate failed to identify oocysts in four of these, and the remaining sample was found to have a number of empty shells with no nuclei evident when examined with DAPI.
Table 2. Cryptosporidium spp. infecting Malawian rural and urban <5-year-olds
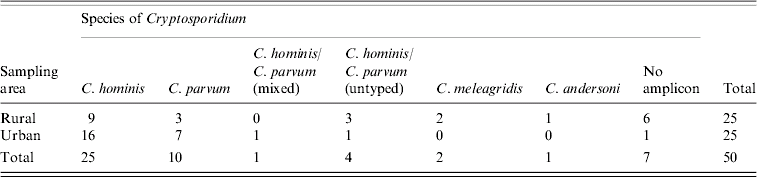
No significant difference in the number of cases infected with different species was seen between the rural and urban areas (χ2 =0·012, 1 d.f., P>0·5) however, the range of species present in Chikwawa was more diverse. Both rural and urban cases were identified primarily in the Malawian rainy season (rainy season 8·1% vs. dry season 3·7%, χ2=6·9, 1 d.f., P<0·01). The median age of cases was 11 months (IQR=9·5) and the median age of participants was 10 months (IQR=11). Eighty-five percent of cases were aged <2 years, and all cases were aged <4 years. Fifty-two percent of cases were between 7 and 12 months old indicating a significantly higher number of cases in that age group (χ2 =22·1, 7 d.f., P<0·005). Age was not an influencing factor with regard to species distribution in cases. With the exception that both our C. meleagridis cases were female, gender was not significantly associated with either cryptosporidiosis or specific Cryptosporidium spp. causing infection. Due to the low numbers of C. meleagridis cases found in this study gender is not thought to be significant.
The sensitivity of the molecular methods used for detecting Cryptosporidium spp. is summarized in Table 3. N18S rRNA proved to be the most consistent, confirming 100% of cases. The STN-COWP and the N-COWP assays did not amplify DNA as consistently as the N18S rRNA gene locus assay [Reference Nichols and Smith22], however, it discriminates between C. hominis and C. parvum. The N18S rRNA gene assay of Xiao et al. [Reference Xiao26] can differentiate between C. parvum and C. hominis, however, it is less sensitive than the N18S rRNA [Reference Nichols and Smith22] and failed to amplify the four samples that were negative by the STN-COWP (Table 3). Species identity was not possible with four samples using this COWP gene locus.
Table 3. Sensitivity of molecular methods for detecting Cryptosporidium species in stools from Malawian rural and urban <5-year-olds

PCR–RFLP, Polymerase chain reaction–restriction fragment length polymorphism; STN-COWP, single tube nested PCR amplification of the Cryptosporidium oocyst wall protein; N-COWP, nested PCR amplification of the Cryptosporidium oocyst wall protein; N-Xiao, nested PCR amplification of 18S rRNA gene loci as described by Xiao et al. [Reference Xiao26]; n.a., not applicable.
* In each case one sample which appeared negative was shown to be inhibitory by the use of the internal control.
† Concentration of PCR product too low to produce visible RFLP pattern.
DISCUSSION
Very few studies have been conducted in sub-Saharan Africa to assess the species of Cryptosporidium present in a population. Previously, microscopical studies for diagnosing cryptosporidiosis in developing countries have assumed that the infecting species were C. parvum and C. hominis. More recently, studies in Kenya, Uganda and Malawi have assessed the species types and have identified C. parvum, C. hominis, C. muris and C. meleagridis [Reference Leav10–Reference Tumwine13]. Despite an insufficient difference in oocyst size and morphology to be able to differentiate the species, this study has shown that three different species were present in children aged <5 years. Each species has different implications for the source of infection, subsequent risk assessment and epidemiology. It is interesting to note that C. hominis is mostly associated with outbreaks of cryptosporidiosis in developed countries with C. parvum being the main cause of sporadic cases [Reference McLaughlin18]. However, in this and other studies in sub-Saharan Africa, the dominant species has consistently been C. hominis in sporadic cases thereby indicating the increased potential for anthroponotic transmission in this environment.
Microscopic examination
mZN generated the lowest number of oocyst-positive samples, and, in this study, led to misdiagnosis and under-diagnosis of infection. The number of positive samples diagnosed with mZN increased as oocyst density increased (44% detection with 0/1+ sample, 100% detection with 3+ samples) as previously described [Reference Baxby and Blundell27–Reference Morgan29]. AP and IF were better at detecting oocyst-positive samples, but neither can be relied on solely for maximizing the detection threshold. The hierarchy for detecting oocysts tinctorially on slides was AP>IF>mZN (Table 1).
Oocysts in 14 samples [C. hominis (n=3), C. parvum (n=1), C. hominis/parvum (n=1), C. andersoni (n=1), C. meleagridis (n=1), no PCR amplification (n=7)] failed to stain with the two FITC-C-mAbs used when labelled on direct smears (un-purified) and after water–ether purification (semi-purified), but Cryptosporidium oocysts/DNA were detected by at least two other methods (mZN, AP, DIC and PCR–RFLP). A significant number of these occurred in samples procured in the rural area (χ2=10, 1 d.f., P<0·005) where a minority of samples may have been subject to temperature fluctuations after deposition and storage prior to collection (maximum 3 days). Not all C. parvum and C. hominis isolates can be stained with any of the commercially available FITC-C-mAbs (R.A.B. Nichols, unpublished observations) and it is likely that these oocysts did not react with the commercial FITC-C-mAbs used in this study.
Molecular techniques
The majority of samples (81%) contained C. hominis or C. parvum oocysts. C. meleagridis was detected in two samples, and is the first description of human infection with C. meleagridis in Malawi. The C. andersoni 18S rDNA amplicon [Reference Nichols24] was detected in one sample, however, insufficient material was available for confirmation. Human infection with C. muris has been reported from Kenya [Reference Gatei30]. C. andersoni has not been previously reported in a human case, however, a putative C. muris-like sample reported in an HIV+ patient in France [Reference Guyot31], had greater sequence similarity to C. andersoni than to C. muris [Reference Palmer32].
Ten oocyst-positive stool samples were lost in transit between Malawi and the SPDL, but the Cryptosporidium spp. infecting these ten cases were identified from oocyst-positive slides, processed for immunofluorescence, and obtained at the time of sample procurement in Malawi. Oocyst density ranged from ± to 3+ and the Cryptosporidium spp. present were detected in all 10 samples using N18S rRNA. A further seven samples, where PCR amplification was unsatisfactory, were assessed using similarly fixed slides and six of these yielded positive results. The overall positivity of PCR–RFLP for species identity from stained slides was 94%. This is similar to that of Amar and colleagues [Reference Amar, Pedraza-Diaz and McLauchlin33, Reference Amar34] who demonstrated an overall sensitivity of 90%, however, in contrast to these workers who stored their samples on slides for up to 8 weeks at room temperature prior to testing, our slide samples were prepared 2 years previously and stored at 15–30°C. Given our problems with sample storage and exposure to temperature fluctuations, we recommend further investigations into the use of oocyst-positive methanol fixed slide samples for species/genotype/subtype identification by PCR, particularly with respect to studies in developing countries where facilities can be limited.
Seven samples did not produce amplicons by PCR, and had no fixed slides from the time of procurement available for further testing. Two samples were shown to be affected by inhibition through the use of an internal control, amplifiable with the same primers [Reference Nichols24]. Inhibition in PCR may be due to a number of factors, not least the diet of the case and effect of polysaccharides associated with consumption of plant material [Reference Monteiro35]. The diet and age of children in which inhibition was noted did not indicate a particular trend in this study (data not shown). No preservatives were used in samples so their inhibitory influence can be ruled out. Microscopic examination by FITC-C-mAbs, DAPI and DIC of the 5 μl of concentrated sample showed that one sample had a number of oocysts, but these were empty, indicating a dearth of DNA to amplify by PCR. Oocysts were not found in the remaining four samples when examined by FITC-C-mAbs, DAPI or DIC. Loss of sporozoite DNA might have occurred during long-term storage without preservative [Reference Johnson23]. The effect of this loss may also have been compounded by the ability of the water–ether concentration technique to concentrate oocysts causing shells and DNA to be lost to waste. Supernatant recovered from samples prior to water–ether concentration and stored at 4°C was subjected to freeze–thaw extraction and PCR amplification for these four samples failed to yield a positive result.
PCR techniques used differed in their efficiency to amplify target DNA, and amplification of the 18S rRNA gene using a two-step method [Reference Nichols, Campbell and Smith21, Reference Johnson23] was the only method found to give 100% positive results for all samples where amplification was anticipated. The inefficiency of the direct compared to the 2SN18S rRNA method was also noted by Nichols et al. [Reference Nichols, Campbell and Smith21]. Amplification of oocyst DNA using the single tube nested PCR for the COWP gene loci was found to be variable with an overall success rate of 47·5% for C. parvum or C. hominis isolates [Reference Homan25]. There was an 80% correlation between samples which failed to produce an amplicon by the single tube nested method targeting the COWP gene loci and the direct 18S rRNA gene loci method, of which only three samples were shown to have any inhibition. Samples which failed to amplify, or produced an inadequate quantity of amplicon for digestion by STN-COWP, were subjected to a 2SN method targeting the same COWP gene loci. Of these 16 samples, 71% produced an amplicon. Therefore, an overall 80% positive result was achieved in samples containing C. parvum or C. hominis when targeting the COWP gene compared to an overall 100% positive result when using the 2SN method targeting the 18S rRNA gene. The results of this study therefore advocate targeting 18S rRNA which allowed the identification of all species of Cryptosporidium to be followed by a 2SN-PCR targeting the COWP gene to differentiate between C. parvum and C. hominis where necessary.
Urban vs. rural distribution
No studies have previously assessed the difference between rural and urban infection rates, and species diversity of cryptosporidiosis in sub-Saharan Africa. It was anticipated that a higher proportion of zoonotic infections would be present in the rural area due to the free-range nature of farming, and the high occurrence of animal faeces in and around homes. We do know that C. parvum oocysts are present in animal stools in this area (Z. Banda, unpublished observations). However, interestingly, we found the majority of C. parvum cases in our urban setting (70%), where animals are not as likely to be present in close proximity to the home. As C. parvum can also be transmitted from person to person, we cannot exclude the possibility that these infections may have been of human origin. A recent study by Peng et al. [Reference Tumwine13] identified C. hominis and C. parvum as the species causing cryptosporidiosis in Blantyre paediatric patients. In addition they indicated that C. hominis was the highest cause of cryptosporidiosis (95%). Our study indicates that although C. hominis was of higher occurrence in the same sample area (64%), C. parvum also played a significant part by causing 36% of infections in the urban setting.
Although the level of C. parvum was not higher than the C. hominis infection as may have been expected, it was interesting to note the increased diversity of species in cases in the rural setting. Both cases of C. meleagridis were found in this area in female patients aged between 6 and 12 months old. Epidemiological data collected during home visits to these cases did not indicate any risk factors different to those of other cases included in the study (data not shown), therefore it is not possible to determine the source of infection at this time. C. meleagridis has been associated with both immunocompetent and immunosuppressed cases [Reference Gatei11, Reference Gatei30, Reference Pedraza-Diaz, Amar and McLauchlin36]. However, the proportion of positive cases in this study is higher than that estimated in the developed world (1%) [Reference Pedraza-Diaz37] at 4% which may mean that the infection is more common in this area due to immunosuppression attributable to the high levels of malnutrition and HIV in this population. As C. meleagridis was only identified in the rural population it may also signify the greater risk of infection due to closer contact with livestock which live within, and in close proximity to households. Animal stools sampled in close proximity to the home of cases, and examined by microscopy failed to identify Cryptosporidium oocysts and could, therefore, not be attributed as the cause of infection (data not shown).
Seasonal distribution
A relationship is evident between the presence of Cryptosporidium spp. and annual rainfall with 70% of cases occurring during the rainy season.
Previous studies have shown correlations between the commencement of the rainy season and an increase in cases of Cryptosporidium spp. in sub-Saharan African countries [Reference Nchito38, Reference Steele, Gove and Meewes39]. Although few studies have identified specific sources of infection for cases, this increase may be attributed to a number of factors which occur in the rainy season. First, oocysts present in both animal- and human-derived faeces may be washed from surrounding areas into drinking water sources. In Malawi, this is compounded by the use of unprotected water sources in the rainy season (Chikwawa 40%, Blantyre 18%), including rivers and unprotected wells, in conjunction with the lack of sanitary facilities (Chikwawa 60%, Blantyre 35%, without sanitary facilities) [2]. In addition, an increase in surface water levels, which provide a source for animals to drink from and children to play in, should also be considered. In high density housing areas (urban), both the close proximity of houses and the lack sanitary facilities may contribute to the rapid spread of infection through poor hygiene and human waste disposal practices. This is supported by the higher number of C. hominis infections, which are primarily associated with person-to-person transmission, in the urban area. Rural incidence between the seasons was not found to be significantly different, compared to that of the urban area. This may be attributed to the higher level of poor sanitation and unprotected water sources in this district, in conjunction with the increased potential for zoonotic spread of infection.
Age distribution
The majority of cases occurred in the 0–24 months age group (88%), and may, in part, reflect the high percentage of children from this age group that were sampled (79%), which may in turn reflect the vulnerability of children in this age group to contract diarrhoeal illness. Fifty-two percent of cases occurred in children aged between 7 and 12 months. Results indicate that there is a significantly higher risk of contracting cryptosporidiosis in that age group (χ2=22·1, 7 d.f., P<0·005). The incidence of all species of Cryptosporidium identified in this study were higher in this age group than any other (C. hominis 40%, C. parvum 30%, C. meleagridis 100%, mixed 100%, C. parvum/C. hominis 50%). This general trend was also noted in a previous Kenyan study [Reference Simwa40], and may be attributed to the reduction in assisted immunity from breastfeeding at that age. Partial immunity from previous exposure may then be protecting the higher age groups from an equivalent rate of infection. This is a phenomenon that requires further seroepidemiological investigation and should be the subject of a further cohort study.
ACKNOWLEDGEMENTS
The authors acknowledge the parents and guardians of the study participants, the staff at the Department of Environmental Health, University of Malawi, QECH and Chikwawa District Hospital for their invaluable assistance in the collection and processing of samples. Funding in part is acknowledged from The Carnegie Trust for the Universities of Scotland, The British Council, Lilongwe, Faculty of Engineering and Malawi Millennium Project, University of Strathclyde, and the Royal Environmental Health Institute for Scotland.
DECLARATION OF INTEREST
None.





