Non-syndromic permanent dentition agenesis is clearly a multifactorial trait, determined by both genetic and environmental factors (Brook, Reference Brook2009). The paired domain box gene 9 (PAX9) and the muscle segment homeodomain-homeobox1 (MSX1) have 16 and 11 identified mutations that lead to non-syndromic tooth loss in humans (see http://www.ncbi.nlm.nih.gov/omim — OMIM#167416; OMIM#142893). For several years, we have conducted research on the correlations between genotype and dental agenesis phenotypes involving these genes (Pereira et al., Reference Pereira, Salzano, Mostowska, Trzeciak, Ruiz-Linares, Chies and Bortolini2006; Paixão-Côrtes et al., Reference Paixão-Côrtes, Braga, Salzano, Mundstock, Mundstock and Bortolini2011a, Reference Paixão-Côrtes, Meyer, Pereira, Mazières, Elion, Krishnamoorty and Bortolini2011b). Relevant information about them is provided in Table 1. During our ongoing research we came across a pair of monozygotic (MZ) twins who showed a different pattern of second premolar and third molar agenesis. PAX9 and MSX1 analyses of the twins and their father revealed interesting results.
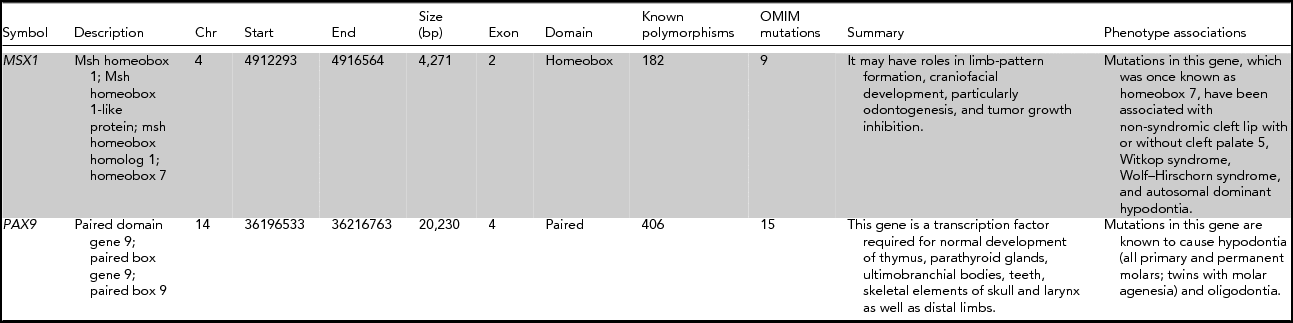
TABLE 1 Information About the Genetic Systems Tested in This Study

Chr = Chromosome; bp = base pair.
Sources of data: the Gene Ontology — AmiGO (http://www.geneontology.org/); Genecards (http://www.genecards.org/index.shtml); Ensemble (http://www.ensembl. org/); OMIM (http://www.ncbi.nlm.nih.gov/sites/entrez?db=omim); and NextBio (http://www.nextbio.com/b/nextbio.nb).
Subjects and Methods
The 12-year-old male twins (patients 1 and 2) discussed in this article are of predominantly European descent, although phenotypic evaluation suggests some level of African ancestry. A detailed clinical and radiographic study of the twins and their father was conducted at the Orthodontic Clinic of the Federal University of Rio Grande do Sul (the mother was not available for study). The three had missing, non-erupted teeth. No other anomaly was found, thereby excluding other syndromes and associated pathologies. We extracted and studied their DNA as described below.
DNA was extracted from saliva using the QIAamp DNAminikit (Qiagen) according to the manufacturer's recommendations.
Amplification of the PAX9 exons 2, 3, 4 and the MSX1 exon 2 (those previously verified to be more prone to variation) was performed in a total volume of 25 μL per reaction, 50 pmol of each primer (Table 2), PCR Master Mix following manufacturer's specifications, and 100 ng of genomic DNA. The components of the reaction were mixed and the sample was placed in a thermal cycler. Polymerase chain reaction (PCR) was performed under the following conditions: 35 cycles of DNA denaturation at 94°C, annealing of primers at 58–59°C, and DNA chain extension (polymerization) at 72°C.
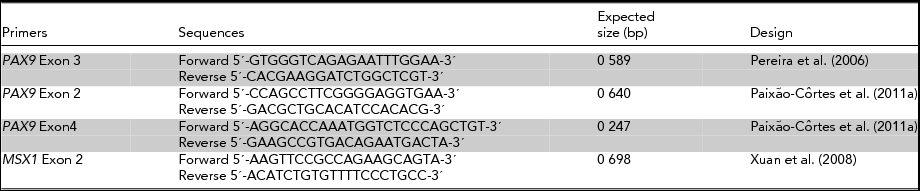
TABLE 2 Primers Utilized in the Amplification Reaction (PCR)

To determine whether the twins were monozygotic or dizygotic (DZ), the AmpFℓ STRIdentifiler™ PCR Amplification kit (Applied Biosystems) was used. The kit tested 15 short tandem repeats (STR) loci: D8S1179, D21S11, D7S820, CSF1PO, D3S1358, TH01, D13S317, D16S539, D2S1338, D19S433, vWA, TPOX, D18S51, D5S818, and FGA. The results were analyzed using the ABI Prism 3130xl Genetic Analyzer (Applied Biosystems) via the GeneMapper Software (Applied Biosystems).
The present research was approved by the Federal University of Rio Grande do Sul Ethics Committee as well as by the Brazilian National Ethics Commission (CONEP protocol no. 9365).
Results and Discussion
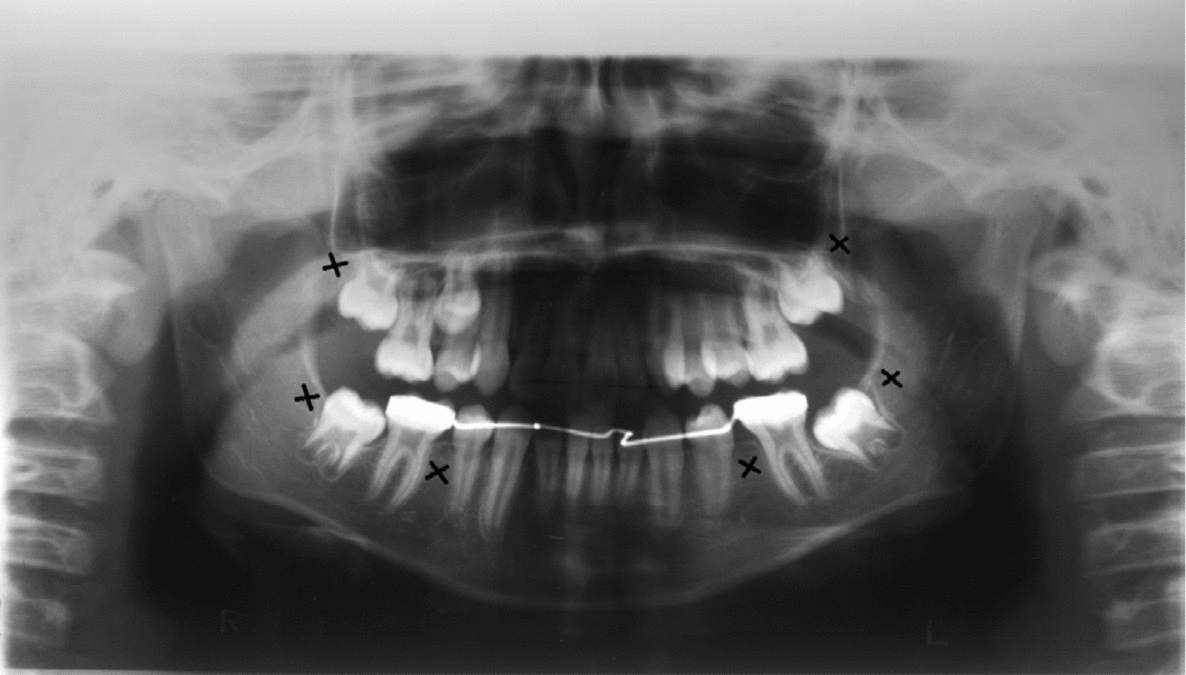
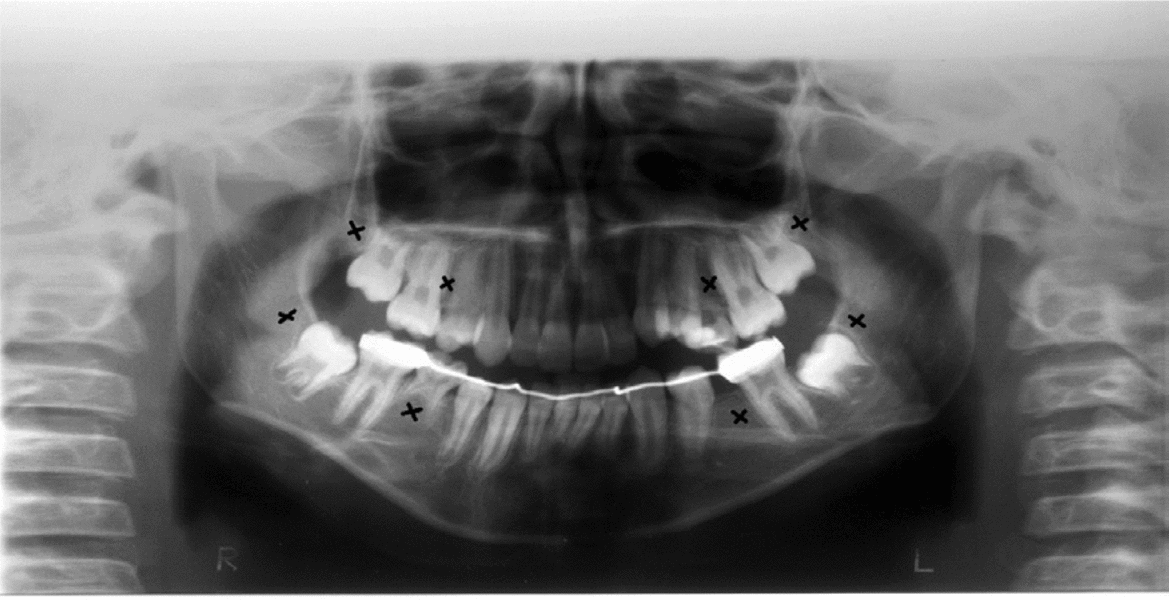
Clinical and radiographic tests revealed that patients 1 and 2 had tooth agenesis, and excluded other causes for the missing teeth, such as dental caries, periodontal disease, or aggressive trauma. Patient 1 had a congenital absence of lower second premolars and upper and lower third molars (Figure 1). In contrast, patient 2 revealed agenesis of upper and lower second premolars, as well as that of upper and lower third molars (Figure 2). Their father also had unilateral agenesis of a lower permanent incisor (data not shown).

FIGURE 1 Panoramic radiograph of patient 1 teeth in 2011; those missing are indicated with an x.

FIGURE 2 Panoramic radiograph of patient 2 teeth in 2011; those missing are indicated with an x.
The twins’ genotypes for the STR markers revealed an identical composition: D8S1179:13/14; D21S11:30/30; D7S820:8/12; CSF1PO:11/12; D3S11358:15/15; THO1:6/9; D13S317:8/12; D16S539:11/12; D2S1338:20/21; D19S433:13/14; vWA:17/17; TPOX:8/8; D18S51:14/17; D5S818:9/11; and FGA:19/25. These results provide a negligible probability value for dizygosis (see, e.g., Yang et al., Reference Yang, Tzeng, Tseng and Huang2006), thereby establishing their MZ nature.
No new mutation in any of the sequenced segments among the three family members was detected. However, in exon 2 of the MSX1 gene we found an already described polymorphism in heterozygosis (rs1095, an exchange of C→T at position 4915839 of chromosome 4; see Supplementary Information in Table S1). Figure 3 presents the sequences of the twins and their father. The mutation was not found in the twins’ father, and therefore must have been inherited from their mother. In an earlier research (Paixão-Côrtes et al., Reference Paixão-Côrtes, Braga, Salzano, Mundstock, Mundstock and Bortolini2011a) we found this mutation in 20% of agenesis patients, while it did not appear among the controls. Third molar agenesis is a common trait found among affected individuals analyzed in the present and aforementioned studies.

FIGURE 3 MSX1 exon 2 chromatograms of (a) the twins (they are identical); and (b) their father.
In the databank of the 1000 Genomes Project, the rs1095 T allele was present among Asians and Latin Americans (15% and 7%, respectively), but not among Africans or Europeans (http://www.1000genomes.org/; 1000 Genomes Project Consortium et al., 2010). We could, therefore, infer a probable Native American origin for this allele among Brazilians and other Latin Americans. The twins’ mother (who had no tooth agenesis) would have received this mutation independent of any agenesis susceptibility. However, due to the fact that the agenesis present in the twins was distinct from that of their father, and considering also its independent presence among other patients (Paixão-Côrtes et al., Reference Paixão-Côrtes, Braga, Salzano, Mundstock, Mundstock and Bortolini2011a), it is not possible to discard the hypothesis that rs1095 T allele can contribute to agenesis tooth manifestation, particularly of third molars.
Paixão-Côrtes et al. (Reference Paixão-Côrtes, Braga, Salzano, Mundstock, Mundstock and Bortolini2011a) identified at least 44 genes that are possibly involved in the dental development network. This and other results suggest that tooth agenesis is influenced by complex population or individual-specific genetic backgrounds (Paixão-Côrtes et al., Reference Paixão-Côrtes, Meyer, Pereira, Mazières, Elion, Krishnamoorty and Bortolini2011b). Table S1 presents the SNPs of the MSX1 and PAX9 genes that have been identified and described up to the present, indicating the degree of variability found among human populations. Furthermore, even among MZ twins, differential expression is not unlikely if they were differently influenced by epigenetic factors. Townsend et al. (Reference Townsend, Richards, Hughes, Pinkerton and Schwerdt2005) found that 21 out of 24 pairs of MZ twins with tooth agenesis also revealed discordant agenesis expression, while Hughes et al. (Reference Hughes, Bockmann, Mihailidis, Bennett, Harris, Seow and Townsend2013) suggested that differences in methylation status at a whole-genome level are related to discordance in the number of teeth found in MZ twins. Indeed, distinct and often undetectable environmental agents may also play a role (Machin, Reference Machin2009).
Acknowledgments
We thank the twins and their father for allowing us to conduct this study. Financial support was granted by Conselho Nacional de Desenvolvimento Científico e Tecnológico and Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (Programa de Apoio a Núcleos de Excelência).
Supplementary Material
To view supplementary material for this article, please visit http://dx.doi.org/10.1017/thg.2013.69.