Introduction
Animal migration and parasitism are intimately linked. From a parasite's perspective, migration of its host provides an opportunity for dispersal to new regions and new pools of potential hosts (Boulinier et al., Reference Boulinier, Kada, Ponchon, Dupraz, Dietrich, Gamble, Bourret, Duriez, Bazire, Tornos, Tveraa, Chambert, Garnier and McCoy2016; Briscoe et al., Reference Briscoe, Nichols, Hartikainen, Knipe, Foster, Green, Okamura and Bass2022). Indeed, parasites that infect migratory host species tend to have larger geographic ranges than parasites that infect only resident host species (de Angeli Dutra et al., Reference de Angeli Dutra, Filion, Fecchio, Braga and Poulin2021a). Host migration has also been hypothesized to promote parasite diversification by facilitating host-switching (Jenkins et al., Reference Jenkins, Thomas, Hellgren and Owens2012), and may influence the evolution of numerous parasite traits such as virulence and lifecycle phenology (Poulin and de Angeli Dutra, Reference Poulin and de Angeli Dutra2021).
From the host perspective, migration can provide an opportunity to escape parasitism if infection prevalence or intensity is locally high year-round and can be avoided by breeding in locales geographically distant from non-breeding sites (Loehle, Reference Loehle1995; Balstad et al., Reference Balstad, Binning, Craft, Zuk and Shaw2021). However, the opposite can be true as well – some host species appear to migrate from regions of low parasite transmission to breeding locations where parasites are frequently acquired (Pulgarín-R et al., Reference Pulgarín-R, Gómez, Bayly, Bensch, FitzGerald, Starkloff, Kirchman, González-Prieto, Hobson, Ungvari-Martin, Skeen, Castaño and Cadena2019). In general, migratory animals appear to host higher parasite species richness than non-migratory animals (Emmenegger et al., Reference Emmenegger, Bauer, Dimitrov, Olano Marin, Zehtindjiev and Hahn2018; Poulin and de Angeli Dutra et al., Reference Poulin and de Angeli Dutra2021). Parasitism can also negatively impact host body condition, preventing a host from becoming physiologically prepared to endure a gruelling migration (Bradley and Altizer, Reference Bradley and Altizer2005; Mages and Dill, Reference Mages and Dill2010; but see Hahn et al., Reference Hahn, Bauer, Dimitrov, Emmenegger, Ivanova, Zehtindjiev and Buttemer2018 for an example of haemosporidian parasites having no detectable impact on avian migratory capacity). Parasitism could also potentially delay migration or impact the ability of a host to properly navigate to their destination (Møller et al., Reference Møller, de Lope and Saino2004; Santiago-Alarcon et al., Reference Santiago-Alarcon, Mettler, Segelbacher and Schaefer2013; Hegemann et al., Reference Hegemann, Alcalde Abril, Sjöberg, Muheim, Alerstam, Nilsson and Hasselquist2018).
Much has been written about the potential for migratory host species to spread parasites across large geographic distances (Altizer et al., Reference Altizer, Bartel and Han2011; Fritzsche McKay and Hoye, Reference Fritzsche McKay and Hoye2016). It is clear from analyses of parasite biogeographic history that parasites can shift transmission areas over evolutionary time (Hellgren et al., Reference Hellgren, Waldenström, Peréz-Tris, Szöll, Si, Hasselquist, Krizanauskiene, Ottosson and Bensch2007; Hoberg and Brooks, Reference Hoberg and Brooks2008), presumably by hitching a ride to new regions using migratory hosts followed by local transmission. However, a lesser-studied phenomenon is the transmission of parasites between local and migratory hosts at stopover sites between the wintering and breeding grounds where migratory animals temporarily pause their movements. Parasite transmission during migration, between passing migrants and local resident individuals, has been speculated on as a potentially important phenomenon that would give parasites numerous opportunities to spread into new communities as their hosts migrate (Figuerola and Green, Reference Figuerola and Green2000; Altizer et al., Reference Altizer, Bartel and Han2011; Ciloglu et al., Reference Ciloglu, Ergen, Inci, Dik, Duzlu, Onder, Yetismis, Bensch, Valkiūnas and Yildirim2020; de Angeli Dutra et al., Reference de Angeli Dutra, Fecchio, Martins Braga and Poulin2021b). However, parasite transmission during migration has been rarely documented, and only in directly transmitted parasites (e.g. viruses in birds, Krauss et al., Reference Krauss, Stallknecht, Negovetich, Niles, Webby and Webster2010; protozoans in butterflies, Satterfield et al., Reference Satterfield, Maerz, Hunter, Flockhart, Hobson, Norris, Streit, de Roode and Altizer2018). An open question is whether vector-mediated parasites are transmitted during host migration, and if so, whether this event occurs routinely (Ishtiaq and Renner, Reference Ishtiaq, Renner, Santiago-Alarcon and Marzal2020).
It is highly difficult to document parasite transmission in action in a natural environment, particularly among highly active and volant hosts such as birds. As a result, the question of whether migratory hosts spread or acquire parasites at stopover locations along their routes has not been directly addressed in most systems. However, there are several ways to test whether parasite transmission between passing migrants that use stopover sites and local hosts is likely to occur without observing the acquisition of parasites in real time. First, we can ask whether the community of parasites that infects migratory hosts overlaps with the parasite community of local hosts. A homogeneous parasite community between migratory and local hosts would suggest frequent parasite transmission between hosts with different migratory strategies, whereas high turnover between them would suggest that there is some barrier to transmission between migratory and local hosts. We can also examine the parasites that are unique to migratory hosts, and test whether they are associated with a climate that is distinct from the stopover site. A parasite carried by a migratory host may not successfully transmit to a local host at a stopover site if the abiotic conditions are not favourable for transmission. Lastly, in the case of vector-borne parasites, we can assess whether the activity of known vector species corresponds with host migratory periods. A lack of temporal overlap between migrant and vector activities would provide strong evidence that transmission between migratory and local hosts is unlikely to occur during stopover.
Blood parasites of birds are an excellent group with which to evaluate transmission between migratory and local hosts at stopover sites. Migration in birds is a well-studied phenomenon, and with the advent of community science initiatives such as eBird (Sullivan et al., Reference Sullivan, Wood, Iliff, Bonney, Fink and Kelling2009) and novel technologies for tracking birds across their annual cycles (McKinnon and Love, Reference McKinnon and Love2018), our understanding of avian migratory routes and stopover duration has greatly accelerated in recent years. Importantly, birds are also hosts to a wide array of easily studied endo- and ectoparasites that can be tracked across the avian annual cycle (Pulgarín-R et al., Reference Pulgarín-R, Gómez, Bayly, Bensch, FitzGerald, Starkloff, Kirchman, González-Prieto, Hobson, Ungvari-Martin, Skeen, Castaño and Cadena2019). Parasites in the order Haemosporida, which includes malaria parasites and their relatives, are one of the best-studied groups of wildlife parasites and are ubiquitous parasites of birds globally (Fecchio et al., Reference Fecchio, Clark, Bell, Skeen, Lutz, De La Torre, Vaughan, Tkach, Schunck, Ferreira, Braga, Lugarini, Wamiti, Dispoto, Galen, Kirchgatter, Sagario, Cueto, González-Acuña, Inumaru, Sato, Schumm, Quillfeldt, Pellegrino, Dharmarajan, Gupta, Robin, Ciloglu, Yildirim, Huang, Chapa-Vargas, Álvarez-Mendizábal, Santiago-Alarcon, Drovetski, Hellgren, Voelker, Ricklefs, Hackett, Collins, Weckstein and Wells2021). The existence of the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009), a centralized repository for avian haemosporidian infection records from tens of thousands of sampled hosts, has made this group of parasites an emerging model for disease ecology.
Although avian haemosporidians have been studied intensively within communities of breeding birds and communities of migrating birds separately, the study of haemosporidians at a single location across multiple seasons is rare (e.g. Huang et al., Reference Huang, Chen, Guocheng, Xia, Luo and Dong2022) and it is even more uncommon to evaluate the potential for transmission of haemosporidians between local and migratory hosts. Here, we tested the hypothesis that haemosporidian transmission between local and migratory birds is infrequent and is limited by climate and vector abundance. We tested this hypothesis by characterizing the haemosporidian communities of local and migratory birds, as well as migratory bird and vector abundances at a temperate stopover site in eastern North America.
Materials and methods
Study site and sample collection
Samples were collected at Rushton Woods Preserve (hereafter referred to as ‘Rushton’), which is 86 acres of protected temperate forest and fields in Newtown Square, Chester County, Pennsylvania (39.984°N, −75.486°W). Sampling occurred between 12 April and 4 November in 2015, 2016 and 2018, and from 22 May to 31 July in 2019. Bird sampling was performed in accordance with the Drexel University Institutional Animal Care and Use Committee under protocol no. 20689 and the USFWS banding permit no. 23679. Birds were captured using mist nets and banded with uniquely numbered USFWS bands. We collected up to 70 μL of blood from each bird by puncturing the brachial vein with a 29-gauge needle. We used this blood to make 2 blood smears on microslides and then froze the remaining blood in liquid nitrogen to preserve DNA. The blood smears were air dried and fixed in methanol. Upon returning to the lab, the slides were stained for 50 min using a 10% Giemsa stain, databased and deposited in the Academy of Natural Sciences of Drexel University bird blood film collection. The collected blood smears were not examined for the current study.
We sought to compare haemosporidian communities between avian hosts that breed at Rushton (‘local breeding birds’) and those that use Rushton temporarily during their annual migrations (‘passing migrants’). We further classified local breeding birds as ‘residents’ that are not known to exhibit migratory behaviour, and migratory species that reproduce at Rushton (‘breeding migrants’). In contrast, passing migrants were classified as either migratory species that are not known to reproduce at Rushton (‘complete passing migrants’), or individuals of migratory species that are known to breed at Rushton, but were using Rushton as a stopover site at the time of sampling (‘non-breeding migrants’). To distinguish between ‘breeding migrants’ and ‘non-breeding migrants’, which can consist of individuals from the same bird species, we used a combination of date cutoffs, evidence of reproductive behaviour (e.g. cloacal protuberance or brood patch) and evidence of site fidelity from banding records. A detailed description of the data sources that we used to make this distinction is given in the Supplementary Methods.
Molecular methods
DNA was extracted from the blood samples using the Qiagen DNeasy 96 Blood & Tissue Kit (Qiagen, Germantown, MD, USA) protocol. Each sample was screened for avian haemosporidian parasites of the genera Plasmodium, Haemoproteus (including the subgenus Parahaemoproteus) and Leucocytozoon using a standard cytochrome b (cytb) barcoding approach following Carlson et al. ( Reference Carlson, Proudfoot, Gentile, Dispoto and Weckstein2018). The resulting consensus sequences were used to identify unique haemosporidian cytb haplotypes, which we refer to as ‘genetic lineages’ following the standard in avian haemosporidian research (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009). The resulting consensus sequences were either assigned to genetic lineages already present in the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009), or were given new lineage names using MalAvi naming standards (first 3 letters of the host genus and species followed by a unique number). Chromatograms with overlapping traces in at least 1 nucleotide position were identified as mixed infections with multiple haemosporidian lineages and parsed apart manually following Starkloff and Galen (Reference Starkloff and Galen2023).
As we were interested in species-level differences in haemosporidian communities, we used the following framework to identify lineages that reflect putative haemosporidian species (rather than intraspecific cytb variants that would inflate community alpha diversity). Any rare (5 or fewer occurrences) lineage that differed from a more abundant lineage by just 1 base pair was ‘lumped’ into the abundant lineage for analysis. This decision was justified by previous studies showing that haemosporidian lineages that differ by 1 base pair often share nuclear genotypes (Haemoproteus: Bensch et al., Reference Bensch, Pérez-Tris, Waldenström and Hellgren2004; Leucocytozoon: Galen et al., Reference Galen, Nunes, Sweet and Perkins2018; Plasmodium: Hellgren et al., Reference Hellgren, Atkinson, Bensch, Albayrak, Dimitrov, Ewen, Kim, Lima, Martin, Palinauskas, Ricklefs, Sehgal, Valkiūnas, Tsuda and Marzal2015).
We identified haemosporidian lineages that are transmitted in North America as those sampled either in host species that do not exhibit migratory behaviour, or in hatch-year birds that had never left North America at the time that they were sampled. Haemosporidians were determined to be transmitted at Rushton specifically if the parasite was found to infect resident host species or hatch-year individuals of migratory species that breed at Rushton and were sampled between 1 June and 10 August.
Phylogenetic analyses
Using BEAST2 (Bouckaert et al., Reference Bouckaert, Heled, Kühnert, Vaughan, Wu, Xie, Suchard, Rambaut and Drummond2014), we estimated a Bayesian phylogeny for all haemosporidian cytb lineages that we detected. We identified the best-fit model of evolution as GTR + I + G using jModelTest2 (Darriba et al., Reference Darriba, Taboada, Doallo and Posada2012), and implemented a strict clock and a Yule speciation model. We ran the analysis for 10 million generations, sampling every 1000 generations. We summarized the posterior distribution of topologies using TreeAnnotator, discarding the first 10% as burn-in to estimate the maximum clade credibility tree. For avian host species, a distribution of 100 phylogenies for all host species that were sampled was downloaded from www.birdtree.org (Jetz et al., Reference Jetz, Thomas, Joy, Hartmann and Mooers2012) using the ‘Ericson all species’ option. A majority clade credibility tree was estimated from this distribution using TreeFinder.
Haemosporidian beta diversity and ecological traits
We characterized the beta diversity of haemosporidian communities infecting local breeding birds and passing migrants at Rushton using R (version 4.1.1, R Core Team, 2022) and the package betapart (Baselga and Orme, Reference Baselga and Orme2012) with the function ‘beta.pair’ to estimate total beta diversity and its turnover (replacement of species) and nestedness (the degree to which 1 community is a subset of another) components using the Jaccard index. We also measured haemosporidian beta diversity among hosts in different migratory categories using the unweighted form of the UniFrac distance (Lozupone and Knight, Reference Lozupone and Knight2005), which we used to incorporate the phylogenetic distances among parasite lineages within communities. We calculated UniFrac distances using the ‘UniFrac’ function in the phyloseq package (McMurdie and Holmes, Reference McMurdie and Holmes2013). To provide more geographic context to the comparison of the haemosporidian communities of birds of different migratory categories at Rushton, we also tested whether the haemosporidian community of passing migrants was more similar to birds that breed at Rushton than to haemosporidians from high-latitude bird communities in North America. For this analysis, we used the previously described beta diversity methods (Jaccard index and UniFrac distance) to compare the data from Rushton to community data from a survey of avian haemosporidians in breeding birds in 6 sites in central Alaska (Galen et al., Reference Galen, Speer and Perkins2019), which is one of the only community-wide survey of avian haemosporidians from a high-latitude location in North America.
In addition, we tested whether local breeding birds and passing migrants at Rushton had significantly different haemosporidian communities. We constructed a host–parasite interaction matrix with hosts as the rows and parasites as the columns, and used a binary variable of ‘host breeds at Rushton’ and ‘host uses Rushton as a stopover site’ as the independent variable. We used the ‘adonis2’ function in the vegan package (Oksanen et al., Reference Oksanen, Simpson, Blanchet, Kindt, Legendre, Minchin, O'Hara, Solymos, Stevens, Szoecs, Wagner, Barbour, Bedward, Bolker, Borcard, Carvalho, Chirico, Caceres, Durand, Evangelista, FitzJohn, Friendly, Furneaux, Hannigan, Hill, Lahti, McGlinn, Ouellette, Cunha, Smith, Stier, Braak and Weedon2022) to conduct a permutational analysis of variance (PERMANOVA) using 999 permutations. We conducted PERMANOVA using 3 beta diversity metrics: the Jaccard index, which uses binary presence/absence data, Bray–Curtis distance, which take into account the abundances of species (here parasites) across different sites (here host species) and both weighted and unweighted UniFrac distances.
Next, we tested for differences in geographic range size and host specificity among haemosporidian lineages that were either exclusive to local breeding birds, exclusive to passing migrants or were shared between the 2 groups. To quantify geographic range size, we used the geosphere package in R (Hijmans, Reference Hijmans2022) to calculate the area of a polygon defined by the geographic coordinates of the sampling locations in North America for each lineage that we detected using the ‘areaPolygon’ function. Coordinates were extracted from the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009). To test for differences in host specificity, we used a phylogenetic approach with the ‘ses.mpd’ function in the picante R package (Kembel et al., Reference Kembel, Cowan, Helmus, Cornwell, Morlon, Ackerly, Blomberg and Webb2010) following the parameters described in Galen et al. (Reference Galen, Ray, Henry and Weckstein2022). The input for this analysis was a host–parasite interaction matrix (parasites as rows, hosts as columns) constructed from host–parasite associations obtained from the ‘hosts and sites’ table in the MalAvi database (accessed 26 July 2023) that was filtered to include only haemosporidian cytb lineages that we sampled. We used phylogenetic analysis of variance (ANOVA) to test for trait differences among haemosporidians that belonged to different host migratory categories using the ‘phylANOVA’ function from the phytools package (Revell, Reference Revell2012).
Lastly, we tested whether haemosporidian lineages that were carried by migratory birds and are transmitted in North America outside of Rushton were associated with a significantly different climate than haemosporidians that we confirmed were transmitted at Rushton. Out of the 52 lineages that we detected at Rushton with confirmed North America transmission, 16 were found to be transmitted outside of Rushton (but not at Rushton), 23 were found to be transmitted at Rushton and 13 lineages did not have additional sampling sites in North America and so were not included in this analysis. We again queried records of the target haemosporidians from the ‘hosts and sites’ table in MalAvi for which sampling coordinates were available. We downloaded temperature variables from the MERRAclim dataset at a 2.5 arc-minute scale (Vega et al., Reference Vega, Pertierra and Olalla-Tárraga2017), and then extracted the temperature variables at each locality in R by creating a combined list of all variables using the velox package (version 0.2.1, Hunziker, Reference Hunziker2021) and extracting the temperature variables by converting all coordinates to spatial points in sp (version 1.5-0, Pebesma and Bivand, Reference Pebesma and Bivand2005; Bivand et al., Reference Bivand, Pebesma and Gómez-Rubio2013) and using the ‘extract_points’ function in sp. We specifically focused on MERRAclim variable BIO10, which corresponds to the mean temperature of the warmest quarter, rather than all bioclimatic variables collectively because we had an a priori expectation from previous research that temperature is particularly influential for haemosporidian sporogonic development in the vector (Valkiūnas, Reference Valkiūnas2004), and can potentially negatively impact transmission (Platonova and Palinauskas, Reference Platonova and Palinauskas2021). We used phylogenetic ANOVA as above to test whether haemosporidians found in migratory birds and not transmitted at Rushton are associated with significantly lower BIO10 values than haemosporidians known to be transmitted at Rushton.
Temporal associations among bird and vector abundances
To characterize the potential for vectors to transmit haemosporidians during migratory stopover periods at Rushton, we used publicly available databases of bird and mosquito abundances in southeastern Pennsylvania. Note that because no publicly available database of abundance values exists for vectors in the families Simuliidae and Ceratopogonidae, the vectors of Leucocytozoon and Haemoproteus, respectively, this analysis only concerns temporal overlap between migratory birds and the mosquito (Culicidae) vectors of Plasmodium.
We used the ebirdst R package (Strimas-Mackey et al., Reference Strimas-Mackey, Ligocki, Auer and Fink2023) to generate estimated counts of migratory bird species that we sampled at Rushton across each week of the year. The counts estimated by ebirdst represent the expected counts of a bird species on a 1 h, 1 km checklist submitted to eBird during an optimal observation period, and are estimated for 2022 (Fink et al., Reference Fink, Auer, Johnston, Strimas-Mackey, Ligocki, Robinson, Hochachka, Jaromczyk, Crowley, Dunham, Stillman, Davies, Rodewald, Ruiz-Gutierrez and Wood2023; Strimas-Mackey et al., Reference Strimas-Mackey, Ligocki, Auer and Fink2023). We also used a publicly available database of mosquito counts from West Nile virus surveys across the state of Pennsylvania (Pennsylvania Department of Environmental Protection, accessed through https://files.dep.state.pa.us/Water/WNV/MosquitoTestingData/ in October 2023). This database includes exact counts of every species of mosquito that was trapped between the months of April and October across 14 901 sampling sites in Pennsylvania between 2016 and 2022. We restricted this dataset to only include: (1) mosquito counts from traps set in Chester County, Pennsylvania (the county in which Rushton is located), (2) mosquitoes sampled from gravid traps that collect mosquitoes that have putatively already fed on a host, which reflects the proportion of a mosquito population that is potentially involved in disease transmission and (3) mosquito species that have been confirmed as vectors of avian malaria parasites based on the review by Santiago-Alarcon et al. (Reference Santiago-Alarcon, Palinauskas and Schaefer2012). We summarized cumulative counts of mosquito species for each week of the sampling season, and corrected for differential sampling efforts across weeks by randomly extracting the smallest number of independent sampling events that occurred during any week of the year (95 sampling events), without replacement. We repeated this subsampling protocol 1000 times and used the average number of mosquitos counted per week from the subsampling procedure for analysis.
Using the estimated weekly counts for migratory bird species and mosquitoes for weeks 18 through 39 of the annual calendar (corresponding approximately to the last week of April through the last week of September), we used cross-correlation analysis to test whether periods of migratory bird and mosquito abundance are decoupled over time. Rather than assess the abundance of all migratory bird species at Rushton, we focused on migratory species that we sampled in the 3 avian families that made up the overwhelming majority (93%) of migrant individuals that were sampled at Rushton: Parulidae, Passerellidae and Turdidae. We used the ‘cc.test’ function from the testcorr package (Dalla et al., Reference Dalla and Phillips2021) to test for the correlation between migratory bird and mosquito abundances over time.
Results
Haemosporidian abundance and diversity
We sampled 1454 birds of 63 species between 2015 and 2019. We restricted this dataset to the 1404 samples of birds in the order Passeriformes due to low sample sizes of non-passerine hosts. Haemosporidian prevalence in individual host species varied widely (Fig. 1). Among residents, prevalence varied from 0% (Sitta carolinensis) to over 90% (Cardinalis cardinalis). We observed similar ranges of prevalence in migratory host species: breeding migrants at Rushton varied from 8% prevalence (Spinus tristis) to 90% (Cyanocitta cristata), whereas complete passing migrants varied from 8% (Regulus calendula) to 100% (Setophaga palmarum). Across all samples, the overall haemosporidian prevalence was 63.1%. Plasmodium was the most commonly encountered parasite with a prevalence of 38.6%, followed by Haemoproteus (including Parahaemoproteus) at 22.4% and Leucocytozoon at 16.8%. Coinfections were common, with 21% of all hosts harbouring multiple haemosporidian infections.

Figure 1. Haemosporidian prevalence of Leucocytozoon, Haemoproteus and Plasmodium in bird species that were sampled at Rushton at least 5 times. Each bird species is classified as a resident (does not exhibit migratory behaviour), a breeding migrant/non-breeding migrant (migratory species that both breed at Rushton and pass through as migrants) or a complete passing migrant (are migratory and are not known to breed at Rushton). The combined haemosporidian infection rate is depicted, which equals the average number of haemosporidian infections per individual of a host species. The combined infection rate is shown as the additive prevalences of Leucocytozoon, Haemoproteus and Plasmodium within each host species.
We recorded 91 haemosporidian cytb lineages (20 Haemoproteus, 41 Leucocytozoon, 30 Plasmodium). Out of 1276 individual haemosporidian detections (including coinfections), 54% were of just 5 common lineages: MAFUS02 (Haemoproteus; 204 detections), PADOM11 (Plasmodium; 185 detections), TUMIG03 (Plasmodium; 111 detections), SEIAUR01 (Plasmodium; 102 detections) and GEOTRI09 (Plasmodium; 93 detections). Twenty-seven lineages were novel, though these lineages were typically rare (5 or fewer detections) and were mostly different from a more common lineage by 1 base pair (22 of the 27 lineages). All lineage infection data have been deposited in the MalAvi database (Bensch et al., Reference Bensch, Hellgren and Pérez-Tris2009), and the sequences for novel lineages have been deposited in GenBank (accession numbers OQ503649–OQ503676). After lumping together similar lineages for analysis, we retained 63 lineages (18 Haemoproteus, 25 Leucocytozoon, 20 Plasmodium). We confirmed North American transmission of 52 of these lineages, 26 of which were confirmed as transmitted at Rushton (Supplementary Table 1). We confirmed that the remaining parasite lineages were transmitted in North America outside of Rushton through our detection of parasites in migrating hatch-year birds during autumn (meaning that they had acquired their infections before ever leaving North America), or from previous records deposited in MalAvi of infections of these lineages in non-migratory bird species in North America (Supplementary Table 1).
Haemosporidian community change across migratory host groups
We classified 557 host individuals as local breeding birds at Rushton (130 residents and 427 breeding migrants), and 737 individuals were classified as passing migrants that do not breed at Rushton and instead use Rushton as a stopover site (283 were complete passing migrants, and 454 were non-breeding migrants). We were unable to classify 110 birds as either breeding migrants or non-breeding migrants, as they were sampled at Rushton during a time when some members of the species are breeding locally and some are still passing through as migrants.
Haemosporidian communities were distinct between local breeding birds and passing migrants at Rushton. Jaccard dissimilarity between the 2 groups was driven primarily by turnover, not nestedness (total beta diversity = 0.58, nestedness = 0.09, turnover = 0.49). When breaking down bird species into 4 migratory categories (residents, breeding migrants, non-breeding migrants and complete passing migrants), high Jaccard dissimilarity was seen between resident species and all other migratory categories (Fig. 2A). Again, beta diversity among the 4 migratory categories was driven primarily by turnover, not nestedness (Fig. 2B). Notably, the non-breeding migrant haemosporidian community was more similar to that of complete passing migrants than it was to breeding migrants, despite the fact that non-breeding migrants and breeding migrants contain overlapping host species. We also compared the haemosporidian community of passing migrants and local breeding birds at Rushton to a high-latitude haemosporidian community that was previously sampled in Alaska. Using the Jaccard index (Fig. 2) and unweighted UniFrac distances, we found that the haemosporidians of passing migrants were more similar to haemosporidians sampled in Alaska than they were to haemosporidians sampled in breeding birds at Rushton (Fig. 2). PERMANOVA indicated that the haemosporidian communities in local breeding birds and passing migratory birds were significantly different, no matter the index used to characterize beta diversity (Jaccard index: F = 1.99, P = 0.001; Bray–Curtis index: F = 2.54, P = 0.002; unweighted UniFrac: F = 3.03, P = 0.012; weighted UniFrac: F = 4.08, 0.011).

Figure 2. Haemosporidian beta diversity across avian migratory categories at Rushton. ‘Alaska’ includes the haemosporidian community found in breeding birds in Alaska by Galen et al. (Reference Galen, Speer and Perkins2019), ‘Complete passing migrant’ is the haemosporidian community sampled in hosts at Rushton that are migratory and do not breed at Rushton, ‘Non-breeding migrant’ includes haemosporidians sampled in passing migrants of species that do breed at Rushton, ‘Breeding migrant’ is the haemosporidian community sampled in migratory host species that were breeding at Rushton and ‘Resident’ is the haemosporidian community of local breeding birds at Rushton that are non-migratory. (A) Lower triangle is the Jaccard distance and the upper triangle is the unweighted UniFrac distance. (B) Lower triangle is the turnover component of the Jaccard distance depicted in panel A and the upper triangle is the nestedness component of the Jaccard distance depicted in panel A.
There was no difference in the geographic distribution of haemosporidians that infected local breeding birds only, passing migrants only or were shared between the 2 groups (phylogenetic ANOVA: F = 2.0, P = 0.22). We also found no difference in host specificity among haemosporidians from different avian migratory categories (phylogenetic ANOVA: F = 2.3, P = 0.14).
Haemosporidians that we confirmed were transmitted at Rushton were associated with additional sampling sites that have significantly higher mean temperatures of the warmest quarter than haemosporidians found in passing migrants that are transmitted in North America outside of Rushton (Fig. 3; phylogenetic ANOVA: F = 3.18, P = 0.047).

Figure 3. Haemosporidian lineages detected in passing migrants that are transmitted in North America, but not at Rushton (‘Not Rushton’, shown in yellow), are associated with sampling sites with lower mean temperatures of the warmest quarter (MERRAclim Bio10) than haemosporidians that we confirmed are transmitted at Rushton (‘Rushton’, shown in purple). Mean MERRAclim Bio10 values (which corresponds to the mean temperature of the warmest quarter) are depicted next to each haemosporidian lineages in the haemosporidian phylogeny, and as a boxplot.
Temporal overlap between migratory bird and mosquito abundance
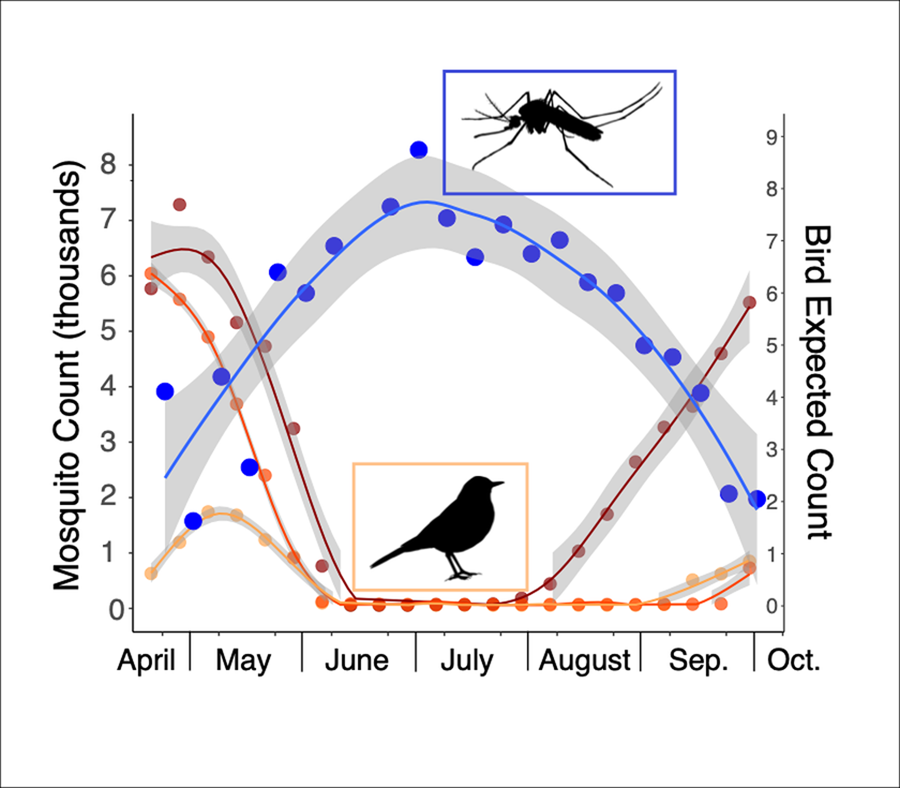
The abundance of all 3 common migratory bird families (Turdidae, Parulidae and Passerellidae) was significantly negatively correlated with mosquito abundance at a time lag of zero, indicating that when bird abundance was high, mosquito abundance was low and vice versa for each week of the study period (Turdidae: t = −2.9, P = 0.003; Parulidae: t = −4.1, P ≤ 0.001; Passerellidae: t = −2.5, 0.014) (Fig. 4). For all 3 bird families, significant positive correlations with mosquito abundance only occurred at time lags of 5 weeks or more, showing a significant temporal offset between when migratory bird abundance and mosquito abundance peak. We repeated this analysis using all mosquito species, including those that have been confirmed as vectors of avian malaria parasites and those that have not, and found virtually identical results (Supplementary Fig. 1). When considering mosquito species individually, we found that a single mosquito species, Culex restuans, exhibited positive correlations with avian abundance (Supplementary Fig. 2, Table 2). Culex restuans was significantly positively correlated with the avian family Turdidae at a time lag of zero (t = 2.4, P = 0.017), though correlations with the families Parulidae and Passerellidae were not significant and all other mosquito species exhibited only negative correlations.

Figure 4. Mosquito and migratory bird abundance over time in southeastern Pennsylvania. (A) Cumulative mosquito count per week based on random sampling of 95 trapping events for annual weeks 18 through 39 between 2016 and 2022. (B) Expected counts of migratory birds in the families Parulidae, Passerellidae and Turdidae at Rushton during weeks 18 through 39 of the annual calendar for the year 2022. Mosquito silhouette was downloaded from www.phylopic.org.
Discussion
Migratory animals clearly influence the global distributions of parasites and are often studied for their potential to spread their parasites throughout their annual cycle (Pulgarín-R et al., Reference Pulgarín-R, Gómez, Bayly, Bensch, FitzGerald, Starkloff, Kirchman, González-Prieto, Hobson, Ungvari-Martin, Skeen, Castaño and Cadena2019; Soares et al., Reference Soares, Latta and Ricklefs2020; Poulin and de Angeli Dutra, Reference Poulin and de Angeli Dutra2021). Of particular interest is the capacity for migratory animals to spread parasites to local hosts during active migration at stopover sites, yet this phenomenon has rarely been investigated in most host–parasite systems. We studied haemosporidians infecting birds that stop briefly at a migratory stopover site to determine the potential for the spread of these parasites between passing migrants and the local bird community. We found multiple lines of evidence to suggest that there is limited potential for transmission of haemosporidians between birds that breed at a temperate forest site and birds that use this site as a temporary stopover location.
Our first goal was to determine whether birds breeding at Rushton and birds using this site for stopover have similar haemosporidian parasite communities. We found that local breeding birds and passing migrants do share a core community of abundant and widespread haemosporidians, with 5 haemosporidian lineages that were shared between local breeding and migrant birds composing over half of all haemosporidian detections: Haemoproteus MAFUS02 and Plasmodium PADOM11, SEIAUR01, TUMIG03 and GEOTRI09. All 5 of these lineages have been recorded dozens of times from multiple sites in North America, and we confirmed that they are transmitted at Rushton. The existence of this group of abundant, widespread haemosporidians suggests that there is potential for the transmission of these lineages between local and passing migrant birds at Rushton. Although transmission of these abundant haemosporidians during migratory stopover is not expected to lead to disease emergence in communities where these lineages already occur, there is still risk of the introduction of these abundant parasites to novel communities by migratory birds, particularly due to vagrancy (Cohen et al., Reference Cohen, Auckland, Marra and Hamer2015).
However, when considering all haemosporidian lineages, multiple beta diversity metrics indicated that the haemosporidian communities of local breeding birds at Rushton and passing migrants are significantly different, whether considering presence/absence, abundances or phylogenetic relationships. The existence of a distinct haemosporidian community in passing migrants at our temperate sampling site was expected, given that many of these host species are known to breed in the North American boreal forest. Recent surveys of haemosporidians in North American boreal forest communities have identified high haemosporidian diversity that is largely distinct from temperate sampling sites (Oakgrove et al., Reference Oakgrove, Harrigan, Loiseau, Guers, Seppi and Sehgal2014; Galen et al., Reference Galen, Speer and Perkins2019; Fecchio et al., Reference Fecchio, Bell, Bosholn, Vaughan, Tkach, Lutz, Cueto, Gorosito, González-Acuña, Stromlund, Kvasager, Comiche, Kirchgatter, Pinho, Berv, Anciães, Fontana, Zyskowski, Sampaio, Dispoto, Galen, Weckstein and Clark2020). Indeed, by comparing the haemosporidian community of passing migrants at Rushton to a haemosporidian community from Alaska, we found evidence that passing migrants sampled at Rushton harbour haemosporidians that are characteristic of high-latitude boreal forests in North America. Collectively, these patterns support the inference that haemosporidians, like their avian hosts, are biogeographically structured in North America as has been shown in South America (Fecchio et al., Reference Fecchio, Bell, Pinheiro, Cueto, Gorosito, Lutz, Gaiotti, Paiva, França, Toledo-Lima, Tolentino, Pinho, Tkach, Fontana, Grande, Santillán, Caparroz, Roos, Bessa, Nogueira, Moura, Nolasco, Comiche, Kirchgatter, Guimarães, Dispoto, Marini, Weckstein, Batalha-Filho and Collins2019; McNew et al., Reference McNew, Barrow, Williamson, Galen, Skeen, DuBay, Gaffney, Johnson, Bautista, Ordoñez, Schmitt, Smiley, Valqui, Bates, Hackett and Witt2021).
What factors have allowed migrant birds at Rushton to maintain a distinct haemosporidian community, despite the fact that these birds clearly intermingle with temperate breeding birds during stopover? Our study suggests that there are several important factors that likely limit the transmission of haemosporidians between migratory and local birds at Rushton and other similar temperate stopover sites. First, there appears to be either a direct or an indirect effect of climate on haemosporidian distributions that may limit transmission potential at stopover sites. Haemosporidians found in passing migrants, for which we do not have evidence of transmission at Rushton, were associated with a distinct climate relative to haemosporidians that are transmitted at Rushton. Specifically, the haemosporidians found in passing migrants were generally recorded at sites with lower temperatures during the warmest quarter of the year. In fact, based on current sampling records (e.g. Haemoproteus SPIARB01 and DUNNO01; Leucocytozoon ACAFLA03, PERCAN01 and POEHUD01; Plasmodium BT7), several haemosporidians that we only found in passing migrants appear to have distributions that are restricted to the boreal zone of North America, suggesting the existence of some barrier to transmission south of this region. One variable that likely structures the distributions of North American haemosporidians is the distribution of their vectors. Haemosporidians not only exhibit specificity at the level of the vector family, but some haemosporidians appear to be transmitted by specific vector genera or clades within a vector genus (Santiago-Alarcon et al., Reference Santiago-Alarcon, Palinauskas and Schaefer2012). The lack of overlap in vector communities between boreal and temperate zones in North America is likely an important contributor to the geographic structure of haemosporidian communities, and may act as a limitation to transmission potential during avian stopover.
In addition to the species composition of the available vector community, a related factor that likely limits the potential for haemosporidian transmission during stopover is vector abundance. For the avian families Turdidae, Parulidae and Passerellidae, which make up the majority of migratory birds that stop temporarily at Rushton, migrant abundance is negatively correlated with overall mosquito abundance throughout the year. For all 3 host families, counts of migratory species peak from late April into mid-May, when mosquito abundance is still low (though we note that the abundance of 1 mosquito species, C. restuans, appears to be an exception to this general pattern). By the time mosquito abundance takes off in early June, migratory birds are virtually gone from the study region. Similarly, mosquito abundance in southeastern Pennsylvania declines sharply in mid-September, just as the abundance of the families Turdidae and Passerellidae begin to climb and the abundance of Parulidae is still increasing. Although these patterns do not preclude transmission from occurring, it does suggest that the probability of a successful transmission event between a migratory and local host at Rushton is negatively impacted by mosquito abundance given that it has been documented that Plasmodium transmission increases with vector density (Koella, Reference Koella1991). Furthermore, the brief periods in the spring and autumn when migratory birds are abundant at Rushton may not be ideal for the development of some haemosporidians in the vector. For instance, temperatures below 15°C, which occur regularly at Rushton particularly during the late spring, appear to significantly delay sporozoite development in some Plasmodium and Haemoproteus species and also inhibit vector biting activity (Atkinson et al., Reference Atkinson, Forrester and Greiner1988; LaPointe et al., Reference LaPointe, Goff and Atkinson2010). Unfortunately, we were unable to analyse vector abundance for the simuliid blackflies and Culicoides biting midges, which are the vectors for Leucocytozoon and Haemoproteus, respectively. Although this does limit our inferences regarding transmission potential for Plasmodium, previous surveys in eastern North America suggest that simuliid and Culicoides abundances could also be decoupled from migratory bird abundance at temperate North American sites (simuliids: Adler et al., Reference Adler, Travis, Kim and Masteller1982; Culicoides: Lysyk, Reference Lysyk2006).
There are numerous intriguing avenues for future research into the potential for migratory animals to spread pathogens at stopover locations. One natural extension of this research is to evaluate the temporal overlap between migratory bird and vector abundances across latitudes, as one might hypothesize that the window of migrant-vector overlap is extended at lower latitudes in North America. For example, in subtropical regions of North America, the abundance of some vectors may be positively correlated with migratory bird abundance as the vectors reach seasonal peaks during the brief window in which most migratory birds pass through. Modelling the effect of climate change on the interaction between vectors and migratory hosts has shown that these windows of time during which migrants and vectors can interact may change over time (Hall et al., Reference Hall, Brown and Altizer2016), and so the effects of warming temperatures should be incorporated in future studies. Lastly, from an evolutionary perspective, it is intriguing to consider whether migratory birds in North America have optimized the timing of their migratory pathways to reduce the temporal overlap with vectors. We were also unable to factor the length of time that migrant birds linger at stopover sites into this analysis. It is interesting to consider that if transmission of vector-mediated pathogens occurs at stopover sites, it may be more likely to occur in the autumn due to the influx of naïve hatch-year birds and the tendency for birds migrating in the autumn to engage in longer stopover durations (Nilsson et al., Reference Nilsson, Klaassen and Alerstam2013). Collectively, our understanding of pathogen transmission during migratory stopover is still in its infancy and is deserving of increased focus.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0031182024001239.
Data availability statement
Genetic data for novel haemosporidian lineages generated by this study have been made available on GenBank (accession numbers: OQ503649–OQ503676). Data for all haemosporidian detections have been submitted to the MalAvi database. The data file and R code used for analysis have been included as Supplementary materials. The accession numbers for the blood smears that were produced for this research are included in the Supplementary data file, and the original blood smears are available for study at the Academy of Natural Sciences of Drexel University.
Acknowledgements
We thank the numerous individuals that have helped band birds at Rushton Woods Preserve who facilitated this research, including AR Ciccariello, Blake Goll, Chyna Poor Thunder-Hutcherson, Colleen Farrell, Doris McGovern, Heather Kostic, Jacqueline Garcia, Jessica Shahan, Kaya Gentile, Matthew R. Halley, Moed Gerveni and Todd Alleger. Matt Helwig and Keith Price from the Pennsylvania Department of Environmental Protection played a critical role in accessing the mosquito abundance data, and we thank them for making these data widely available in an easily accessible format.
Author contributions
S. C. G. conceptualized the project, collected samples, analysed data and wrote the first draft of the manuscript. E. O. collected samples and performed MERRAclim analysis. S. R. and M. H. collected samples and performed lab work to detect haemosporidian infections, J. D. performed lab work to detect haemosporidian infections, A. F. and L. K. collected samples and J. D. W. conceptualized the project and made major contributions to the manuscript. All authors read and approved the final manuscript.
Financial support
This work was supported in part by NSF Postdoctoral Research Fellowship in Biology 1811806 to S. C. G., a Drexel University Summer Research Award to J. D. W. and the Academy of Natural Sciences Ornithology Department Endowment. Undergraduate interns on the project were supported in part by the Jim Stewart Memorial Fund of the Academy of Natural Sciences of Drexel University.
Competing interests
None.
Ethical standards
Bird sampling was performed in accordance with the Drexel University Institutional Animal Care and Use Committee under protocol no. 20689 and the USFWS banding permit no. 23679.