Global seafood consumption, currently estimated to be 100 million tonnes, is expected to increase by another 65 million tonnes by the year 2030. Aquaculture already makes a significant contribution to this seafood supply, and this is growing at an annual rate close to 6 %( 1 ). Most farmed teleost fish are adapted to use amino acids as the preferred energy source over carbohydrates and thus require high levels of dietary proteins (300–600 g/kg)( Reference Cowey 2 ), with fishmeal being the major protein source in aquaculture. The sustainability of this practice, which requires large inputs of wild fish for feed preparation, has been questioned( Reference Naylor, Hardy and Bureau 3 ). Thus, the replacement of fishmeal as the major protein source with proteins of plant origin is a major objective for sustainable aquaculture in the future. Progress has been made on the replacement of fishmeal with a number of alternative protein sources of plant origin (oil seeds, cereal proteins and pulses)( Reference Carter and Hauler 4 – Reference Gatlin, Barrows and Brown 7 ). However, the replacement of fishmeal with plant proteins is often limited by methionine concentrations. In this context, supplementation of plant-based diets with synthetic methionine has been shown to optimise the nutritional value of these diets containing alternative proteins, with dietary methionine having a significant effect on growth performance and protein deposition in different fish species( Reference Rodehutscord, Jacobs and Pack 8 – Reference Sveier, Nordås and Berge 11 ). Given that muscle protein accretion is the major aim of aquaculture, the optimisation of dietary methionine supply requires precise knowledge of the role of this sulphur amino acid, especially in the muscle.
For optimising methionine nutrition, the different roles of this amino acid need to be considered. Methionine is an indispensable amino acid for the normal growth of most animals. This sulphur amino acid is a component of tissue proteins and therefore serves as a building block for protein synthesis. Decreased growth in chicks or mammals fed methionine-free diets has thus been reported to be primarily caused by lower rates of whole-body protein synthesis associated with lower mRNA translational efficiency( Reference Kino and Okumura 12 ). These findings are in agreement with the specific and well-known role of methionine in mRNA translation as the primary amino acid needed to initiate protein synthesis( Reference Bradshaw, Brickey and Walker 13 ). However, there is evidence that this amino acid not only affects muscle growth as a substrate for protein synthesis but may also modulate several intracellular signalling pathways involved in the regulation of mRNA translation and the two major proteolytic pathways (ubiquitin–proteasome and autophagy-lysosomal). More precisely, methionine availability has been reported to regulate the target of rapamycin complex 1 (TORC1) signalling pathway and protein synthesis in avian QM7 myoblasts( Reference Métayer-Coustard, Mameri and Seiliez 14 ) and the expression of the proteasomal gene F-box protein 32 (Fbx32, also known as atrogin-1 or MAFbx) in quail QT6 cells( Reference Tesseraud, Métayer-Coustard and Boussaid 15 ). Similarly, the eukaryotic translation initiation factor 2α/activating transcription factor 4 (eIF2α/ATF4) pathway has recently been shown to direct an autophagy gene transcriptional programme in response to methionine starvation in mouse embryonic fibroblast cells( Reference B'chir, Maurin and Carraro 16 ). This amino acid could thus act as a regulator of muscle growth in the same manner as certain hormones (e.g. insulin and insulin-like growth factor 1) in accordance with the concept of ‘nutrient signal’ developed over the last 10 years for amino acids such as leucine( Reference Anthony, Yoshizawa and Anthony 17 – Reference Yoshizawa 21 ).
Several studies have demonstrated that supplementation of methionine in diets rich in plant proteins can improve the growth response of many fish species( Reference Rodehutscord, Jacobs and Pack 8 , Reference Takagi, Shimeno and Hosokawa 22 , Reference Gaylord and Barrows 23 ). To gain more knowledge on the influence of methionine intake on growth and nutrient accretion, several groups have investigated the effect of graded concentrations of methionine in the diet on the hepatic metabolism of fish( Reference Cowey, Cho and Sivak 24 – Reference Craig and Moon 29 ). The results of our previous study on rainbow trout hepatocytes have revealed that methionine availability controls the activation of the TORC1 intracellular signalling pathway as well as the expression of several metabolism-related genes in this species( Reference Lansard, Panserat and Plagnes-Juan 27 ). These data suggest that the signalling role of methionine is well conserved between lower and higher vertebrates. However, until now, no data are available on the specific role of this amino acid in the molecular control of muscle growth in any fish species. As muscle protein synthesis rates are low in fish despite the high efficiency of protein deposition, it is considered essential to gain full insight into the protein degradation pathways( Reference Kaushik and Seiliez 30 ). The aim of the present study was to determine the effect of diets that differ in methionine concentrations on the mechanisms involved in muscle protein turnover in rainbow trout (Oncorhynchus mykiss). More specifically, we investigated the effect of dietary methionine deficiency or excess on several main factors involved in the regulation of mRNA translation and the two major muscle proteolytic pathways (ubiquitin–proteasome and autophagy-lysosomal).
Materials and methods
The present experiment was carried out in accordance with the EU legal frameworks, specifically those related to the protection of animals used for scientific purposes (i.e. Directive 2010/63/EU), and under the French legislation governing the ethical treatment of animals (Decret no. 2001-464, 29 May 2001). The investigators carrying out the experiment had ‘level 1’ or ‘level 2’ certification, bestowed by the Direction Départementale des Services Vétérinaires (French veterinary services) to carry out animal experiments (Institut National de la Recherche Agronomique (INRA) 2002-36, 14 April 2002).
Feeding trial and experimental procedures
Sexually immature rainbow trout (O. mykiss) having a mean initial weight of 40 g were reared in our experimental fish farm facilities (INRA, Donzacq, France) in a flow-through rearing system supplied with natural spring water (17°C) under natural photoperiod during the months of December–January. The fish were distributed into nine circular tanks (300 litres; three tanks per diet; thirty eight fish/tank). Triplicate groups of trout were fed for 6 weeks with one of the three isonitrogenous extruded diets manufactured in our experimental facilities of Donzacq, France (Table 1). The diets differed only in dietary methionine concentrations: deficient (DEF); adequate (ADQ); excess (EXC). The adequate concentration (0·82 g methionine or 1·25 g total sulphur amino acids methionine+cysteine per 100 g diet) corresponds to the requirement value for rainbow trout (NRC, 2011). The concentration of cysteine was maintained constant at 0·45 g cysteine per 100 g of diet. Each diet was distributed by hand to visual satiation (two meals/d, at 08.00–09.00 and 16.00–17.00 hours), and feed consumption was recorded per tank. To follow growth and feed utilisation, the fish were counted and group-weighed at the start and end of the experiment.
Table 1 Ingredients and analytical composition of the diets
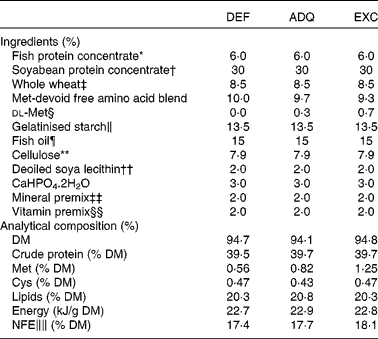
DEF, methionine deficient; ADQ, methionine adequate; EXC, methionine excess; NFE, N-free extract.
* CPSP-90 (Sopropeche).
† Estril 75(Sopropeche).
‡ Sudouest aliment (Pomarez).
§ Evonik.
∥ Roquette.
¶ Sopropeche.
** Rettenmaier & Sohne.
†† Louis François.
‡‡ Mineral premix (g or mg/kg diet): calcium carbonate (40 % Ca), 2·15 g; magnesium oxide (60 % Mg), 1·24 g; ferric citrate, 0·2 g; potassium iodide (75 % I), 0·4 mg; zinc sulphate (36 % Zn), 0·4 g; copper sulphate (25 % Cu), 0·3 g; manganese sulphate (33 % Mn), 0·3 g; dibasic calcium phosphate (20 % Ca, 18 % P), 5 g; cobalt sulphate, 2 mg; sodium selenite (30 % Se), 3 mg; KCl, 0·9 g; NaCl, 0·4 g (UPAE, INRA).
§§ Vitamin premix (μg or mg/kg diet): dl-α-tocopherol acetate, 60 mg; sodium menadione bisulphate, 5 mg; retinyl acetate, 4·5 mg; dl-cholecalciferol, 375 μg; thiamin, 15 mg; riboflavin, 30 mg; pyridoxine, 15 mg; B12, 0·05 mg; nicotinic acid, 175 mg; folic acid, 500 mg; inositol, 1000 mg; biotin, 2·5 mg; calcium pantothenate, 50 mg; choline chloride, 2000 mg (UPAE, INRA).
∥∥ NFE (carbohydrate): 100 % (crude protein%+crude fat%+crude fibre%+moisture%+ash%).
Individual body mass was calculated as the final biomass divided by the number of fish in each tank. Daily growth coefficient was calculated as 100 × (mean final body mass1/3− mean initial body mass1/3)/d. Daily feed intake was calculated as 100 × the total amount of ingested food (kg) divided by the mean biomass over the experimental period ((initial biomass+final biomass)/2, expressed as kg wet mass) and the number of days. Feed efficiency was estimated as the gain in total biomass ((final biomass − initial biomass) (kg wet mass)) divided by the amount of ingested DM (kg DM) (Table 2).
Table 2 Growth performance of trout fed the experimental diets for 6 weeks (Mean values with their standard errors; n 3 tanks per diet)
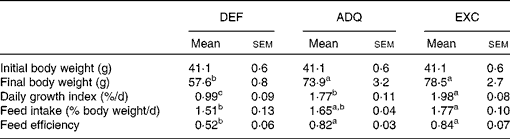
DEF, methionine deficient; ADQ, methionine adequate; EXC, methionine excess.
a,b,cMean values with unlike superscript letters were significantly different among the three dietary groups (P< 0·05; one-way ANOVA followed by the Student–Newman–Keuls multiple-comparison test).
At the end of the feeding trial, three fish were anaesthetised with benzocaine solution (30 mg/l water) and sampled from each tank at 4, 8 and 16 h after feeding the last meal. The fish were killed by a sharp blow to the head. The gut content of the sampled fish was systematically checked to ensure that the fish sampled had effectively consumed the diet. A sample of laterodorsal white muscle collected from each fish was dissected, snap-frozen in liquid N2 and then stored at − 80°C before further mRNA and protein analyses.
Chemical composition of the diets
The chemical composition of the diets was determined as follows: (1) DM was determined after drying at 105°C for 24 h; (2) protein content (N content × 6·25) was determined using the Kjeldahl method after acid digestion; (3) fat content was determined by petroleum diethyl ether extraction (Soxtherm); (4) gross energy content was determined in an adiabatic bomb calorimeter (IKA). Dietary amino acid concentrations were determined after hydrolysis with 6 m-HCl for 24 h at 110°C and quantified by ion exchange chromatography with post-column derivatisation with ninhydrin( Reference Llames and Fontaine 31 ).
Protein extraction and Western blot analyses
The muscle samples of fish sampled at 4 h after feeding the last meal (300 mg) were homogenised on ice using an ULTRA-TURRAX homogeniser (IMLAB Sarl). During homogenisation, the samples were kept in 2 ml of buffer containing 150 mm-NaCl, 10 mm-Tris, 1 mm-ethylene glycol tetraacetic acid, 1 mm-EDTA, 100 mm-sodium fluoride, 4 mm-sodium pyrophosphate, 2 mm-sodium orthovanadate, 1 % Triton X-100, 0·5 % Nonidet P-40/IGEPAL and a protease inhibitor cocktail (Roche). The homogenates were centrifuged at 1000 g for 15 min at 4°C, and the supernatants were then centrifuged for 30 min at 20 000 g . The resulting supernatants (n 6 per time point) were stored at − 80°C. Protein concentrations were determined using the Bradford reagent method( Reference Bradford 32 ). The muscle samples (20 μg) were subjected to SDS–PAGE and Western blot analyses using the appropriate antibodies: anti-phosho S6 (Ser235/Ser236, #4856; Cell Signaling Technologies); anti-carboxyl terminal S6 (#2217; Cell Signaling Technologies); eIF4E (4E)-binding protein 1 (4E-BP1, Thr37/Thr46, #9459; Cell Signaling Technologies); anti-4E-BP1 (#9452; Cell Signaling Technologies); anti-phospho eIF2α (Ser51, #9721; Cell Signaling Technologies); anti-carboxyl terminal eIF2α (#9722; Cell Signaling Technologies); anti-microtubule-associated protein light chain 3B (LC3B, #2775; Cell Signaling Technologies); anti-sequestosome 1 (SQSTM1, #10 117; Santa Cruz); anti-beclin 1 (#3738; Cell Signaling Technologies); anti-β-tubulin (#2146; Cell Signaling Technologies). All primary antibodies used have been shown to cross-react successfully with rainbow trout proteins of interest( Reference Belghit, Panserat and Sadoul 33 – Reference Seiliez, Taty Taty and Bugeon 37 ). For beclin 1, the amino acid sequence of the corresponding protein was monitored in the SIGENAE database (http://www.sigenae.org) to check for the conservation of the antigen sequence with the corresponding sequence from mammals, ensuring a good specificity (79 %) of the mammalian antibody used in the analysis of the samples. After washing, the membranes were incubated with an IRDye IR secondary antibody (LI-COR, Inc., Biotechnology). The bands were visualised by IR fluorescence using the Odyssey® Imaging System (LI-COR, Inc.) and quantified using the Odyssey Infrared Imaging System software (version 1.2; Application Software).
Analysis of mRNA levels: quantitative RT-PCR
The analysis of mRNA levels was carried out using the white muscle samples of fish sampled at 4, 8 and 16 h after feeding the last meal. Total RNA was extracted using TRIzol Reagent (Invitrogen) according to the manufacturer's recommendations. Using the SuperScript III RNaseH–RT kit (Invitrogen) with random primers (Promega), 1 μg of the resulting total RNA (n 6 per time point) was reverse-transcribed into complementary DNA (cDNA) according to the manufacturer's instructions. Details regarding the primer sequences used in the quantitative real-time RT-PCR assays, as well as the protocol conditions of the assays, have been published previously( Reference Seiliez, Panserat and Lansard 35 , Reference Seiliez, Gabillard and Riflade 36 , Reference Seiliez, Gutierrez and Salmerón 38 ). Primers of the SQSTM1, UV radiation resistance-associated gene protein (Uvrag), Bcl-2/adenovirus E1B 19 kDa-interacting protein 3 (Bnip3), mitochondrial ubiquitin ligase activator of NF-κB 1 (Mul1), zinc finger protein 216 (ZNF216) and tripartite motif-containing 32 (Trim32) genes were newly designed using Primer3 software, as described previously( Reference Seiliez, Gutierrez and Salmerón 38 ). To confirm the specificity of the newly developed RT-PCR assay, the amplicon was purified and sequenced (Beckman-Coulter Genomics). The primers used in the real-time RT-PCR assays are listed in Table 3. Quantitative RT-PCR assays were carried out on the Roche LightCycler 480 System (Roche Diagnostics). The assays were carried out using a reaction mix of 6 μl per sample, each of which contained 2 μl of diluted cDNA template, 0·24 μl of each primer (10 μm), 3 μl of LightCycler 480 SYBRH Green I Master Mix and 0·52 μl of DNase/RNase-free water (5 PRIME GmbH). The PCR protocol was initiated at 95°C for 10 min for the initial denaturation of the cDNA and the activation of the hot-start Taq-polymerase, followed by forty-five cycles of a three-step amplification programme (15 s at 95°C, 10 s at 60–64°C and 15 s at 72°C), according to the primer set used (Table 3). Melting curves were systematically monitored (temperature gradient at 1·1°C/10 s from 65 to 94°C) at the end of the last amplification cycle to confirm the specificity of the amplification reaction. Each PCR assay included replicate samples (duplicate of reverse transcription and PCR amplification, respectively) and negative controls (RT- and cDNA template-free samples, respectively). For the expression analysis of mRNA, relative quantification of target gene expression was done using the ΔC T method described by Pfaffl et al. ( Reference Pfaffl, Horgan and Dempfle 39 ). The relative gene expression value of elongation factor 1α (EF1α) was used for the normalisation of the measured expression values of the target mRNA, and it was found to not change significantly over time (data not shown). In all cases, PCR efficiency was measured by the slope of a standard curve using serial dilutions of cDNA. In all cases, PCR efficiency values ranged between 1·8 and 2·2.
Table 3 Sequences of the primer pairs used in the quantitative real-time RT-PCR assays

atg12 l, Autophagy-related 12-like; atg4b, autophagy-related 4b; Uvrag, UV radiation resistance-associated gene; SQSTM1, sequestosome 1; Mul1, mitochondrial ubiquitin ligase activator of NF-κB 1; Bnip3, Bcl-2/adenovirus E1B 19 kDa-interacting protein 3; Fbx32, F-box protein 32; MuRF1, -2, -3, muscle RING finger 1, -2, -3; ZNF216, zinc finger protein 216; Trim32, tripartite motif-containing 32; EF1α, elongation factor 1α.
Statistical analyses
Results are expressed as means with their standard errors (n 6). Statistical analyses of growth performance and protein phosphorylation were carried out using one-way ANOVA to detect significant differences. Gene expression data were analysed using two-way ANOVA. The Newman–Keuls multiple-range test was used to compare means in case of a significant effect (P< 0·05).
Results
Growth performance, feed intake and feed efficiency
The feed intake of fish fed the ADQ diet was similar to that of fish fed the DEF and EXC diets, but it differed slightly between the latter two groups (Table 2). As expected, at the end of the 6-week feeding trial, the final body weight as well as the daily growth index was lower in fish fed the DEF diet than in those fed the ADQ and EXC diets (Table 2). Also, feed efficiency was higher in fish fed the ADQ and EXC diets than in those fed the DEF diet (Table 2).
Muscle S6, 4E binding protein 1 and eukaryotic translation initiation factor 2α phosphorylation status
We investigated the effect of feeding the three experimental diets with graded concentrations of dietary methionine on the phosphorylation of three translation initiation factors (S6, 4E-BP1 and eIF2α) in the muscle of trout sampled 4 h after feeding the last meal. As shown in Fig. 1(A), the phosphorylation of S6 in fish fed the DEF diet was significantly reduced compared with that in fish fed the EXC diet, whereas intermediate results were obtained for fish fed the ADQ diet. In contrast, the phosphorylation of 4E-BP1 was not different among the three dietary groups (Fig. 1(B)). Finally, the phosphorylation of eIF2α was significantly lower in fish fed the EXC diet than in those fed the DEF diet, but it did not differ between the fish fed the ADQ diet and those fed the EXC and DEF diets (Fig. 1(C)).

Fig. 1 Results of the Western blot analysis of (A) S6, (B) 4E binding protein 1 (4E-BP1) and (C) eukaryotic translation initiation factor 2α (eIF2α) protein phosphorylation (p) in the muscle of trout fed the methionine-deficient (DEF), -adequate (ADQ) and -excess (EXC) diets and sampled 4 h after feeding the last meal. Total protein (20 μg per lane) was loaded on the gel. Western blot analysis was carried out on six individual samples per treatment, and a representative blot is shown. Graphs show the ratio of the amount of the phosphorylated protein:the total amount of the targeted protein. Values are means (n 6), with their standard errors represented by vertical bars. a,bMean values with unlike letters were significantly different among the three dietary groups (P< 0·05; one-way ANOVA followed by the Student–Newman–Keuls multiple-comparison test).
Levels of the muscle autophagy-related markers microtubule-associated protein light chain 3-II, sequestosome 1 and beclin 1
We monitored the levels of LC3-II as well as those of SQSTM1 and beclin 1 in the skeletal muscle of trout fed the three experimental diets and sampled 4 h after feeding the last meal. To date, the detection of processed LC3-II by Western blot analysis has been the most reliable method for autophagy detection. As shown in Fig. 2(A), the ratio of LC3-II:β-tubulin reached significantly lower levels in fish fed the EXC diet compared with that in fish fed the DEF diet. In addition to LC3-II, SQSTM1 and beclin 1 can also be used as autophagy markers. There was no significant difference in the ratio of SQSTM1:β-tubulin among the dietary groups (Fig. 2(B)). Similar to the results obtained for LC3-II, the ratio of beclin 1:β-tubulin reached significantly lower levels in fish fed the EXC diet compared with that in fish fed the DEF diet (Fig. 2(C)).

Fig. 2 Results of the Western blot analysis of (A) microtubule-associated protein light chain 3-II (LC3-II), (B) sequestosome 1 (SQSTM1) and (C) beclin 1 proteins in the muscle of trout fed the methionine-deficient (DEF), -adequate (ADQ) and -excess (EXC) diets and sampled 4 h after feeding the last meal. Total protein (20 μg per lane) was loaded on the gel. Western blot analysis was carried out on six individual samples per treatment, and a representative blot is shown. Graphs show the ratio of the targeted protein:β-tubulin used as a loading control. Values are means (n 6), with their standard errors represented by vertical bars. a,bMean values with unlike letters were significantly different among the three dietary groups (P< 0·05; one-way ANOVA followed by the Student–Newman–Keuls multiple-comparison test).
mRNA levels of autophagy-related genes in the skeletal muscle of trout fed diets differing in methionine concentrations and sampled 4, 8 and 16 h after feeding the last meal
We monitored the expression levels of several autophagy-related genes (autophagy-related 4b (atg4b), autophagy-related 12-like (atg12 l), Uvrag, SQSTM1, Mul1 and Bnip3) in the muscle of fish fed the three experimental diets. As shown in Fig. 3, the mRNA levels of all the monitored autophagy-related genes were significantly higher in fish fed the DEF diet than in those fed the EXC diet. Intermediate results were obtained for fish fed the ADQ diet, with their mRNA levels being similar to those of fish fed the DEF and/or EXC diets according to the monitored gene. With the exception of Uvrag mRNA, exhibiting a significant increase in its levels between 8 and 16 h after feeding (Fig. 3(C)), no significant differences were observed among the sampling times for any of the genes studied (Fig. 3(A), (B), (D), (E) and (F)).

Fig. 3 Autophagy-related gene expression in the muscle of trout fed the methionine-deficient (DEF), -adequate (ADQ) and -excess (EXC) diets and sampled 4 h (■), 8 h (![]() ) and 16 h (□) after feeding the last meal. The mRNA levels of (A) autophagy-related 12-like (atg12 l), (B) autophagy-related 4b (atg4b), (C) UV radiation resistance-associated gene protein (Uvrag), (D) sequestosome 1 (SQSTM1), (E) mitochondrial E3 ubiquitin protein ligase 1 (Mul1) and (F) Bcl-2/adenovirus E1B 19 kDa-interacting protein 3 (Bnip3) were measured using quantitative real-time RT-PCR assays. Expression values were normalised to those of the elongation factor 1α (EF1α) transcripts. Values are means (n 6), with their standard errors represented by vertical bars. A significant diet effect was observed for atg12 l mRNA (P< 0·001; DEF = ADQ>EXC), atg4b mRNA (P= 0·003; DEF>ADQ = EXC), SQSTM1 mRNA (P< 0·0001; DEF>ADQ = EXC), Mul1 mRNA (P< 0·0001; DEF>ADQ = EXC) and Bnip3 mRNA (P= 0·0039; DEF = ADQ; ADQ = EXC; DEF>EXC) (two-way ANOVA). Significant effects of diet (P= 0·005; DEF = ADQ; ADQ = EXC; DEF>EXC) and time (P= 0·018; 4 h = 8 h; 4 h = 16 h; 16 h>18 h) were observed for Uvrag mRNA (two-way ANOVA).
) and 16 h (□) after feeding the last meal. The mRNA levels of (A) autophagy-related 12-like (atg12 l), (B) autophagy-related 4b (atg4b), (C) UV radiation resistance-associated gene protein (Uvrag), (D) sequestosome 1 (SQSTM1), (E) mitochondrial E3 ubiquitin protein ligase 1 (Mul1) and (F) Bcl-2/adenovirus E1B 19 kDa-interacting protein 3 (Bnip3) were measured using quantitative real-time RT-PCR assays. Expression values were normalised to those of the elongation factor 1α (EF1α) transcripts. Values are means (n 6), with their standard errors represented by vertical bars. A significant diet effect was observed for atg12 l mRNA (P< 0·001; DEF = ADQ>EXC), atg4b mRNA (P= 0·003; DEF>ADQ = EXC), SQSTM1 mRNA (P< 0·0001; DEF>ADQ = EXC), Mul1 mRNA (P< 0·0001; DEF>ADQ = EXC) and Bnip3 mRNA (P= 0·0039; DEF = ADQ; ADQ = EXC; DEF>EXC) (two-way ANOVA). Significant effects of diet (P= 0·005; DEF = ADQ; ADQ = EXC; DEF>EXC) and time (P= 0·018; 4 h = 8 h; 4 h = 16 h; 16 h>18 h) were observed for Uvrag mRNA (two-way ANOVA).
mRNA levels of proteasomal genes in the skeletal muscle of trout fed diets differing in methionine concentration and sampled at 4, 8 and 16 h after feeding the last meal
We monitored the mRNA levels of several proteasome-related genes (Fbx32, muscle RING finger 1 (MuRF1), MuRF2 and MuRF3, ZNF216 and Trim32) in the muscle of trout fed the three experimental diets. With the exception of MuRF1, the expression of which remained the same among the three dietary groups (Fig. 4(B)), the mRNA levels of all the other monitored proteasome-related genes exhibited a significant reduction in the muscle of fish fed the EXC diet compared with those of fish fed the DEF diet (Fig. 4(A) and (C)–(F)). However, the ‘diet’ effect on Fbx32 mRNA levels was evident only at 4 h after feeding, as revealed by the significant ‘diet × time’ interaction and post hoc Newman–Keuls test (Fig. 4(A)). Similar to the results obtained for the autophagy-related genes, intermediate results were obtained for fish fed the ADQ diet, with their mRNA levels being similar to those of fish fed the DEF and/or EXC diets according to the monitored gene.

Fig. 4 Proteasomal gene expression in the muscle of rainbow trout fed the methionine-deficient (DEF), -adequate (ADQ) and -excess (EXC) diets and sampled 4 h (■), 8 h (![]() ) and 16 h (□) after feeding the last meal. The mRNA levels of (A) Fbx protein 32 (Fbx32), (B–D) the three muscle-specific RING finger 1 genes (MuRF1, MuRF2 and MuRF3, respectively), (E) zinc finger protein 216 (ZNF216) and (F) tripartite motif-containing protein 32 (Trim32) were measured using quantitative real-time RT-PCR assays. Expression values were normalised to those of the elongation factor 1α (EF1α) transcripts. Values are means (n 6), with their standard errors represented by vertical bars. a,bMean values with unlike letters were significantly different among the three dietary groups (P< 0·05; two-way ANOVA followed by the Student–Newman–Keuls multiple-comparison test). For Fbx32 mRNA, significant effects of diet (P= 0·0097; DEF>ADQ = EXC) and time (P= 2·947 × 10− 5; 4 h>8 h = 16 h) were observed and the diet × time interaction was significant (P= 0·0073). A significant diet effect was observed for MuRF2 mRNA (P= 0·0019; DEF>ADQ = EXC), ZNF216 mRNA (P= 0·0446; DEF = ADQ; ADQ = EXC; DEF>EXC) and Trim32 mRNA (P= 0·004; DEF>ADQ = EXC). Significant effects of diet (P= 0·0013; DEF = ADQ>EXC) and time (P= 0·0216; 4 h = 16 h>8 h) were observed for MuRF3 mRNA.
) and 16 h (□) after feeding the last meal. The mRNA levels of (A) Fbx protein 32 (Fbx32), (B–D) the three muscle-specific RING finger 1 genes (MuRF1, MuRF2 and MuRF3, respectively), (E) zinc finger protein 216 (ZNF216) and (F) tripartite motif-containing protein 32 (Trim32) were measured using quantitative real-time RT-PCR assays. Expression values were normalised to those of the elongation factor 1α (EF1α) transcripts. Values are means (n 6), with their standard errors represented by vertical bars. a,bMean values with unlike letters were significantly different among the three dietary groups (P< 0·05; two-way ANOVA followed by the Student–Newman–Keuls multiple-comparison test). For Fbx32 mRNA, significant effects of diet (P= 0·0097; DEF>ADQ = EXC) and time (P= 2·947 × 10− 5; 4 h>8 h = 16 h) were observed and the diet × time interaction was significant (P= 0·0073). A significant diet effect was observed for MuRF2 mRNA (P= 0·0019; DEF>ADQ = EXC), ZNF216 mRNA (P= 0·0446; DEF = ADQ; ADQ = EXC; DEF>EXC) and Trim32 mRNA (P= 0·004; DEF>ADQ = EXC). Significant effects of diet (P= 0·0013; DEF = ADQ>EXC) and time (P= 0·0216; 4 h = 16 h>8 h) were observed for MuRF3 mRNA.
Discussion
The replacement of fishmeal with alternative protein sources remains a major thrust area of research for sustainable aquaculture and much has been accomplished in reducing the utilisations levels of fishmeal in most species( Reference Lim, Webster and Lee 40 ). However, the use of plant protein ingredients often necessitates the addition of one or several crystalline amino acids to meet the essential amino acid requirements. Methionine is the most limiting essential amino acid in oil-seed-derived feedstuffs, such as soya-derived protein sources. Supplementation of diets containing these protein sources with synthetic methionine has been shown to optimise the nutritional value of these diets( Reference Rodehutscord, Jacobs and Pack 8 – Reference Sveier, Nordås and Berge 11 , Reference Mambrini, Roem and Carvèdi 41 ), but excess dietary methionine has also been reported to affect growth performance and composition( Reference Jackson and Capper 42 ). In this context, the precise determination of the molecular basis of the growth reduction induced by methionine-imbalanced diets represents an essential objective for aquaculture. In the present study, we investigated the effect of feeding diets containing different concentrations of methionine on several main factors involved in the regulation of muscle protein turnover in rainbow trout.
We found that rainbow trout fed the amino acid-imbalanced diets (DEF and EXC) did not increase their feed intake (compared with those fed the ADQ diet) to compensate for the inadequate amount of methionine supplied by this diet. At the end of the 6-week feeding trial, the final body weight as well as the feed efficiency was lower in fish fed the DEF diet than in those fed the ADQ and EXC diets. These results confirm the previously described effects of methionine availability on growth and feed utilisation in fish( Reference Rumsey, Page and Scott 10 , Reference Cowey, Cho and Sivak 24 , Reference Mambrini, Roem and Carvèdi 41 , Reference Kim, Kayes and Amundson 43 ) and provide a relevant design for studying the role of methionine in muscle growth-related functions in rainbow trout.
We first investigated the phosphorylation of three translation initiation factors known to play a prominent role in muscle protein synthesis, namely S6, 4E-BP1 and eIF2α( Reference Kimball, Shantz and Horetsky 44 , Reference Kimball and Jefferson 45 ). The results of this analysis revealed that dietary methionine concentrations could modify the phosphorylation of S6. These results are consistent with those of a previous study on avian QM7 myoblasts showing that the phosphorylation of this protein is closely related to the concentration of methionine in the culture medium( Reference Métayer-Coustard, Mameri and Seiliez 14 ). According to these authors, this effect of methionine on the phosphorylation of S6 is mediated by the TORC1 signalling pathway, in agreement with the concept of ‘nutrient signal’ developed for amino acids such as leucine( Reference Anthony, Yoshizawa and Anthony 17 – Reference Yoshizawa 21 ). However, we failed to detect any significant change in the phosphorylation status of the TORC1 downstream effector, the 4E-BP1, between the dietary groups. Such a discrepancy between the phosphorylation levels of these two proteins (S6 and 4E-BP1) has already been observed in the liver of rats in response to low dietary methionine intake( Reference Sikalidis, Mazor and Kang 46 ) and highlights the complexity of the signalling network involved in the regulation of the activation of these translation initiation factors. In addition, we found that the phosphorylation of eIF2α was significantly induced in the muscle of trout fed the DEF diet than in the muscle of trout fed the EXC diet. This result is in line with previous findings demonstrating that growing rats fed a sulphur amino acid-deficient diet exhibit increased phosphorylation of eIF2α in the liver( Reference Sikalidis and Stipanuk 47 ). This effect is consistent with the activation of a stress-induced eIF2α kinase, presumably GCN2 (general control non-derepressible 2), in response to sulphur amino acid deprivation. However, in contrast to previous studies on the role of GCN2 and/or eIF2α phosphorylation in response to amino acid deprivation( Reference Anthony, McDaniel and Byerley 48 ), the present study used diets that were limiting in, but not deficient in, methionine, an essential amino acid. Thus, the present results extend the observations to the situation in which the diet is only deficient in one essential amino acid (i.e. methionine), to an amino acid pattern that is more representative of that in diets consumed by humans and animals, and to a diet that supports growth. Overall, our results showed that rainbow trout respond to a methionine-deficient diet by affecting the phosphorylation of two main translation initiation factors (S6 and eIF2α) in the skeletal white muscle, probably leading to reduced growth and feed utilisation.
As muscle growth is determined by the balance between the rate of protein synthesis and that of protein degradation, we also attempted to address the question whether dietary methionine concentrations affect the key components of proteolysis in the muscle of trout. The ubiquitin–proteasome and autophagy-lysosomal pathways are the two major intracellular proteolytic pathways in the skeletal muscle of vertebrates( Reference Sandri 49 ). Macroautophagy (hereafter referred to as autophagy) functions as an important catabolic mechanism by mediating the turnover of intracellular organelles and protein complexes through a lysosome-dependent degradative pathway( Reference Chen and Klionsky 50 , Reference Cuervo 51 ). In mammals, two main mechanisms have been identified for the induction of autophagy under stress conditions. The first is a rapid and transient transcription-independent induction mediated by TORC1 and AMP-activated protein kinase complexes( Reference Kim, Kundu and Viollet 52 ). During this induction, LC3B is converted from a slower-migrating unconjugated form (LC3-I) to a faster-migrating lipid-conjugated form (LC3-II)( Reference Mizushima and Yoshimori 53 , Reference Klionsky, Abdalla and Abeliovich 54 ). Our previous findings have revealed that the composition of macronutrients (protein:carbohydrate ratio) affects the feeding-mediated short-term inhibition of autophagy in the muscle of rainbow trout( Reference Belghit, Panserat and Sadoul 33 ). More precisely, we demonstrated that LC3-II levels in the muscle of rainbow trout are inhibited by re-feeding only when the proportion of dietary proteins increased at the expense of carbohydrates. In the present study, the levels of LC3-II as well as those of beclin 1 (another essential factor in autophagy interactome) were significantly increased by methionine deficiency in the diet, indicating that the formation of autophagosomes is affected not only by the protein:carbohydrate ratio in the diet but also by the profile of amino acids in the protein fraction. The autophagy-inhibitory effect of amino acids (especially leucine) has been well established in cell-culture experiments( Reference B'chir, Maurin and Carraro 16 , Reference Mordier, Deval and Béchet 55 , Reference Lorin, Tol and Bauvy 56 ). However, to our knowledge, no findings have been published to date on the in vivo effects of dietary deficiency of an essential amino acid on this degradative system.
The second regulatory mechanism of autophagy induction is a slower one requiring gene expression( Reference Mammucari, Milan and Romanello 57 , Reference Füllgrabe, Klionsky and Joseph 58 ). In the present study, we found that the mRNA levels of all the monitored autophagy-related genes were significantly higher in fish fed the DEF diet than in those fed the EXC diet. This result is consistent with our previous findings showing that amino acid availability regulates the expression of several autophagy-related genes in trout myocytes( Reference Seiliez, Gabillard and Riflade 36 ). Similarly, a recent study on mouse embryonic fibroblast cells has reported the up-regulation of a large number of autophagy genes in response to leucine starvation and identified the GCN2/eIF2α pathway as a central regulator of this effect( Reference B'chir, Maurin and Carraro 16 ). Interestingly, the present results reveal that even in vivo the deficiency of methionine in the diet is also associated with an increase in eIF2α phosphorylation, making possible the involvement of this signalling pathway in the observed induction of autophagy-related genes in fish fed the DEF diet. Collectively, the present results reveal that the deficiency of methionine in the diet leads not only to the induction of the formation of autophagosomes in the muscle (revealed by LC3-II levels) but also to the increase of the mRNA levels of several autophagy genes. At physiological levels, such an induction of the expression of these genes in prolonged stress conditions has been reported to be critical to replenish key autophagy-related proteins that are destroyed during prolonged activation of autophagy( Reference Sandri 49 ).
The ubiquitin–proteasome pathway is another important proteolytic pathway that has long been considered to be the primary pathway involved in muscle atrophy( Reference Attaix, Taillandier, Bittar and Rivett 59 – Reference Kumamoto, Fujimoto and Ito 62 ). Protein degradation through this pathway relies on the selective attachment of ubiquitin molecules to the protein substrate by E3 ubiquitin protein ligases. Following polyubiquitination, the targeted proteins are recognised and degraded by the 26S proteasome. Among the genes encoding the E3 ubiquitin ligases, Fbx32 and MuRF1 have been studied in depth and shown to play a key role in the control of skeletal muscle mass( Reference Glass 63 ). These genes are both muscle specific and up-regulated during muscle atrophy( Reference Bonaldo and Sandri 64 , Reference Schiaffino, Dyar and Ciciliot 65 ). In the present study, the mRNA levels of Fbx32 as well as those of two MuRF paralogues (MuRF2 and MuRF3) were significantly higher in the muscle of trout fed the DEF diet than in that of fish fed the EXC diet. These findings are in agreement with the results of a previous study on avian QT6 cells, showing that methionine availability controls the levels of Fbx32 mRNA through a TORC1-dependent mechanism( Reference Tesseraud, Métayer-Coustard and Boussaid 15 ). In contrast, we failed to detect any difference in the mRNA levels of MuRF1 between the dietary groups, in accordance with the results of previous studies indicating differences in the in vivo regulation of Fbx32 and MuRF1 expression( Reference Tesseraud, Bouvarel and Collin 66 , Reference Frost, Nystrom and Jefferson 67 ). Recently, additional proteins involved in muscle protein ubiquitination and proteasome-dependent breakdown have been identified( Reference Sandri 68 ). A recent paper has reported that Trim32 is a crucial E3 ligase for the degradation of thin filaments (actin, tropomyosin and troponins), α-actinin and desmin( Reference Cohen, Zhai and Gygi 69 ). ZNF216 has also been identified as an important player in the recognition and delivery of ubiquitinated proteins to the proteasome during muscle atrophy( Reference Hishiya, Iemura and Natsume 70 ). In the present study, the expression of these two genes was significantly induced when fish were fed the DEF diet. To our knowledge, the present study is the first to provide evidence that the amino acid composition of the diet can modulate the expression of these two genes. Overall, the present results reveal that methionine deficiency induces the expression of major factors involved in the different steps of ubiquitin–proteasome degradation, from the attachment of ubiquitin molecules to the protein substrate to the recognition and delivery of the ubiquitinated proteins to the proteasome.
In conclusion, we demonstrated that feeding rainbow trout with a diet deficient in methionine for 6 weeks results in the suppression of growth performance and affects the activation and/or expression of several key factors involved in muscle growth. More precisely, methionine deficiency affects the phosphorylation of the two main translation initiation factors S6 and eIF2α and induces the expression of several factors involved in the two major muscle proteolytic pathways (ubiquitin–proteasome and autophagy-lysosomal). Whether such changes in the expression and/or activation of these growth-related factors are induced by methionine limitation is unknown, as these changes may be a direct effect of a deficiency of an essential amino acid or an indirect effect of metabolic and or/hormonal changes induced by such a deficiency. Our findings provide the first-ever evidence of the involvement of dietary methionine in mechanisms regulating the muscle growth of fish under physiological conditions. From a practical aquaculture point of view, the present study provides valuable information on biomarkers associated with muscle growth and sulphur amino acid deficiency, a situation not uncommon in fish fed diets rich in plant-based ingredients.
Acknowledgements
The authors thank Evonik-Degussa Laboratory for the analysis of the amino acid composition of the diets. They also cordially thank C. Biran and C. Prochasson from the INRA for their technical assistance as well as the technical staff at the fish farm (F. Vallée, F. Terrier, A. Lanuque and F. Sandres).
The present study was supported by the European Union 7th Framework project (Project Call Identifier: FP7-KBBE-2011-5, Project no.: 288925, Advanced Research Initiatives for Nutrition & Aquaculture (ARRAINA)). The ARRAINA European project also provided a fellowship to I. B.
The authors' contributions are as follows: I. B., S. S.-C., I. G., S. K., S. P. and I. S. designed the study; I. B., K. D. and A. S. conducted the study; I. B., S. S.-C., I. G., S. P. and I. S. analysed the data; I. B. S. P. and I. S. wrote the article; I. S. had primary responsibility for the final content. All authors read and approved the final manuscript.
None of the authors has any conflicts of interest to declare.









