INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) can cause a range of serious disease syndromes in humans, including diarrhoea, haemorrhagic colitis and life-threatening, haemolytic–uraemic syndrome (HUS) [Reference Madigan and Martinko1]. Transmission of STEC can be foodborne, waterborne or from person to person [Reference Nataro and Kaper2]. Ruminants, in particular cattle, are considered to be the main reservoir of human STEC infections, in particular those caused by serogroup O157 [Reference Chapman3]. However, STEC have also been isolated from other domestic animals, wild mammals and birds [Reference Beutin4–Reference Wallace, Cheasty and Jones6] and Shiga toxin-production genes (stx) have also been found in a range of bacteria in the environment [Reference Muniesa7].
Two types of Shiga toxin, stx1 and stx2 (encoded by stx1 and stx2 genes), are associated with human disease. These toxins vary in their amino-acid sequence [Reference Kaper and O'Brian8], antigenicity, and in their activation and receptor specificity [Reference Schmitt, Meysick and O'Brien9]. E. coli acquire stx genes, and the subsequent ability to produce toxins, following infection with temperate bacteriophages [Reference James10].
The ability of E. coli to adhere to intestinal epithelium is crucial in the colonization of the intestine, and therefore the progression of disease in humans. The protein intimin, encoded by the eae gene, enables intimate attachment of E. coli to intestinal cells [Reference Donnenberg and Kaper11], causing characteristic attaching/effacing lesions [Reference Paton and Paton12]. This attachment also enables Shiga toxins to be injected into the epithelial cytoplasm through a type III secretion system [Reference Kaper, Nataro and Mobley13].
Although wild birds have the potential to play a role both as reservoirs and long-distance vectors of STEC (and therefore stx and eae genes), studies of STEC in wild bird populations have been limited to either surveys for the O157 serogroup of E. coli [Reference Wallace, Cheasty and Jones6, Reference Foster14, Reference Wahlstrom15], or to a limited range of wild bird species [Reference Pedersen16–Reference Kobayashi, Pohjanvirta and Pelkonen18]. The aim of this study was therefore to investigate the prevalence, host range and associated risk factors for the occurrence of STEC and the virulence genes stx1, stx2 and eae in E. coli, in a wider range of British wild bird populations. Stx genes are known to be transmitted horizontally between E. coli by highly mobile bacteriophages [Reference James10], therefore investigating the occurrence and distribution of these genes is of great importance to better understand the complex epidemiology and evolution of STEC.
METHODS
Study design
Serial cross-sectional surveys of wild bird populations throughout northern England were undertaken between July 2004 and October 2006. Samples were collected from a large range of wild bird species including representatives from the majority of wild bird families that are present in the UK for part of (migratory species) if not the entire year (resident species). Wild bird individuals were the basic sampling unit.
Sample sources
Samples were collected from both live and dead wild birds. In the UK, live wild birds can only be caught by persons holding a licence issued by the Department for Environment, Food and Rural Affairs (Defra), through a bird ringing scheme run by the British Trust for Ornithology (BTO). All live wild birds sampled in this study were trapped in collaboration with licensed BTO bird ringers [Reference Redfern and Clark19]. A variety of methods were employed to catch wild birds including mist netting, cannon netting, ground traps, wildfowl round-ups, wildfowl shooting, sampling nestlings in the nest, and environmental sampling.
Dead garden birds were collected as part of a collaborative project between the Institute of Zoology and the University of Liverpool; the Garden Bird Health Initiative (GBHi), a study investigating causes of garden bird mortality [Reference Cunningham20].
Sample collection
Once captured, each live bird was placed in a paper bag [Reference Mulvihill and Leberman21], in which they usually defecated allowing a faecal sample to be collected. A new paper bag was used for each bird. Faecal samples were collected from the bags with sterile cotton-tipped swabs, placed in bacterial transport media (Medical Wire and Equipment Ltd, UK) and were transported to the laboratory in a cool box. If the bird did not defecate in the bag, or the bird was too large to place in a bag, a cloacal swab was taken. Occasionally, faecal samples were collected from the environment with sterile swabs as described above. Faecal samples were also collected from the lower intestine of dead birds during post-mortem examination.
Data collection
Data were recorded in the field about the bird and the site at which the bird had been caught or found. The date, location and habitat type where the bird had been trapped or found dead were recorded as well as the capture method, BTO ring number, species, age and sex.
The age and sex of a bird were determined by looking at a combination of features often specific to each species including plumage characteristics, presence or absence of a moult limit, and the size of the bird. When possible, birds were assigned to an age category using the BTO recommended age codes [Reference Redfern and Clark19]. Depending on the species or time of year, not all of the measurements could be taken.
Isolation of E. coli
About 0·5 g faecal material from each bird, or the cloacal swab were placed in 2 ml of brain-heart infusion broth (LabM, UK) containing 5% glycerol (Sigma-Aldrich, UK) and homogenized by vortexing. Five hundred microlitres of the faecal suspension were added to 4 ml buffered peptone water that was incubated aerobically at 37°C for 24±4 h. A loop of the buffered peptone water was subcultured onto eosin Methylene Blue agar (EMBA; LabM), which was incubated aerobically for 20–24 h at 37°C. Ten colonies from each agar plate with a metallic green morphology typical of E. coli were frozen in 2-ml tubes filled with small beads in glycerol (Pro-Lab Diagnostics, UK) at −80°C until they were required.
Examination for virulence genes
Isolates were screened by polymerase chain reaction (PCR) for the presence of STEC virulence genes eae, stx1 and stx2. DNA was prepared directly from colonies that had been frozen at −80°C. Crude DNA extracts were prepared for each isolate by boiling one bead in 0·5 ml sterile distilled water for 20 min in a 1·7-ml microcentrifuge tube. Cell lysates were kept at 4°C for no longer than 14 days. ‘Reddy mix’ PCR master mix (ABgene) was used for the PCR with a final reaction volume of 25 μl. The final concentrations of the components in the PCR mix were 1·25 U Taq DNA polymerase (ABgene), 75 mm Tris–HCl, 20 mm (NH4)SO4, 1·5 mm MgCl2, 0·01% (v/v) Tween-20, 0·2 mm each of dATP, dCTP, dGTP and dTTP, 10 pmol/μl each primer and 1 μl DNA template. The presence of the intimin gene (eae) was investigated using primer sequences forward (sequence 5′ to 3′); GCTTAGTGCTGGTTTAGGATTG and reverse CCAGTGAACTACCGTCAAAG [Reference Beebakhee22, Reference Yu and Kaper23]. Stx genes were detected using the primers slt1-F, CGCTGTTGTACCTGGAAAGG; slt1-R, CGCTCTGCAATAGGTACTCC [Reference La Ragione24], slt2-F, GCTTCTGCTGTGACAGTGAC; slt2-R, TCCATGACAACGGACAGCAG [Reference La Ragione24]. The PCR reaction was carried out with an initial denaturation step at 94°C for 2 min followed by 25 cycles of amplification (denaturation at 94°C for 1 min, annealing at 62°C for 1 min 30 s and extension at 72°C for 2 min). A strain of E. coli serotype O157, isolated from cattle faecal samples and known to be positive for eae, stx1 and stx2 genes, was used as a positive control for each of the PCR assays. PCR products were separated by gel electrophoresis using a 1·5% agarose gel in Tris–acetate buffer. Ethidium bromide was added to the agarose, and the gel was visualized under ultraviolet light. Product sizes were 625 bp, 250 bp and 190 bp, respectively, for eae, stx1 and stx2.
Descriptive statistics
The prevalence of E. coli and each of the virulence genes, and their 95% confidence intervals (CI) were determined using the frequency command in the statistical software package EpiInfo 2002 (CDC, USA).
Univariate analysis
In all analyses, individual wild birds were the analytical units. Univariate and multivariable analyses were carried out using three different binary dependent variables: (1) the occurrence of E. coli that possessed the stx1 gene; (2) the stx2 gene and (3) the eae gene. Univariate associations between binary explanatory variables and the three dependent variables were investigated by performing univariable χ2 tests or Fisher's exact tests (for expected values <5) in Stata v.8 (Stata Corporation, USA). Univariate relationships between the dependent variables and independent categorical variables were examined using χ2 tests for differences between groups and χ2 tests for trend in Stata v.8. Data from wild bird species from which fewer than 10 individuals were sampled were excluded from the analysis due to small sample size.
Multivariable analysis
Multivariable logistic regression models were built for each dependent variable using maximum-likelihood estimation. The location at which birds were caught or found was included in the models as a random effect to account for the possibility that birds caught at the same location were more likely to be similar to each other than birds caught at different locations. The models were built using the xtlogit command in Stata v.8. After univariable screening, variables showing an association with the outcome variable with a P value of <0·25 were considered for inclusion in the multivariable model [Reference Hosmer and Lemeshow25]. Variables with >10% missing values were not entered into the final models to avoid numerical instability. Simple correlation analysis was performed on independent binary variables. Categorical variables were assessed for correlation using the Cramer's phi-prime test statistic, which was carried out in SPSS v.15.0 (SPSS Inc., USA). Associations were considered statistically significant at a P value ⩽0·05. Where two categorical variables were significantly associated and the Cramer's phi-prime test statistic value was >0·3, the directional measure lambda was calculated to provide an indication of how strong the association between each categorical variable was. Statistically significant associations between two categorical variables with a lambda value of >0·3 were considered to be strongly associated and therefore correlated. Of the correlated variables, those with the strongest association with the outcome variable or those variables that made most biological sense were considered for inclusion in the final model to avoid covariance between predictor variables.
The final models were built manually. Variables were entered individually; a maximum-likelihood test (P<0·05) and the model deviance were used to assess how well parameters improved the fit of the model. Parameters with a term-wise Wald test statistic of P<0·05, and parameters that significantly improved the fit of the model (maximum-likelihood χ2 test statistic P<0·05) were retained in the final model. Once each final model had been built, all explanatory variables were re-entered individually to ensure that there were no more significant improvements.
The final models were evaluated for confounding by observing changes in parameter coefficients during the model-building process. Two-way interaction terms were tested between model parameters. For random-effects multivariable models, the number of quadrature points used in the random-effects estimator was checked for stability using the quadchk function in Stata v.8. Stata uses Gauss–Hermite quadrature approximation when estimating random-effects models. Twelve quadrature points are used by default when models are being built. To determine the soundness of quadrature approximation, the model was re-estimated using different numbers of quadrature points (8 and 16). The log-likelihood and coefficient estimates for the original and re-estimated models were compared and a relative difference of <1% was taken as evidence that the choice of quadrature points used did not significantly affect the outcome. To assess the percentage of variation that was attributable to the level of the location at which the bird was sampled, the intra-class correlation coefficient was calculated using a latent variable approach [Reference Goldstein, Browne and Rasbash26]. In models where the random effect was not statistically significant, a logistic regression model was built and the fit of the final model was assessed by examining the model sensitivity and specificity [area under the receiver operating characteristic (ROC) curve] and the Hosmer–Lemeshow test statistic.
RESULTS
Descriptive epidemiology
Faecal samples were collected from 2084 wild birds of 99 different species from 167 locations between July 2004 and October 2006. The overall prevalence of presumptive E. coli isolated from faecal samples was 50·8% (95% CI 48·6–52·9%), isolated from 94 bird species. Of those samples from which E. coli was isolated, the prevalence of stx1, stx2 and eae genes was 1·5% (95% CI 0·9–2·5), 7·9% (95% CI 6·4–9·8) and 4·9% (95% CI 3·7–6·4), respectively. The stx1 genes were isolated from 12 wild bird species, stx2 genes from 30 species and eae genes from 27 species of wild bird. There were significant differences in the monthly prevalence of E. coli (P<0·001), both virulence genes stx1 (P=0·02) and stx2 (P<0·001) but not eae (P=0·17). In the majority of samples, eae, stx1 or stx2 genes were detected individually; however, in 0·4% (95% CI 0·2–1·0) of samples both stx1 and eae genes were detected, and in 0·2% (95% CI 0·05–0·7) of samples both stx2 and eae genes were detected. It should be noted that multiple virulence genes were not necessarily derived from the same E. coli isolate as a pool of 10 E. coli isolates that were cultured from each faecal sample were screened by PCR. E. coli was most prevalent during January, June and October (Fig. 1). The stx1 gene was detected most frequently in E. coli isolated in June whereas stx2 showed peaks of occurrence in February, June, September and October (Fig. 2).
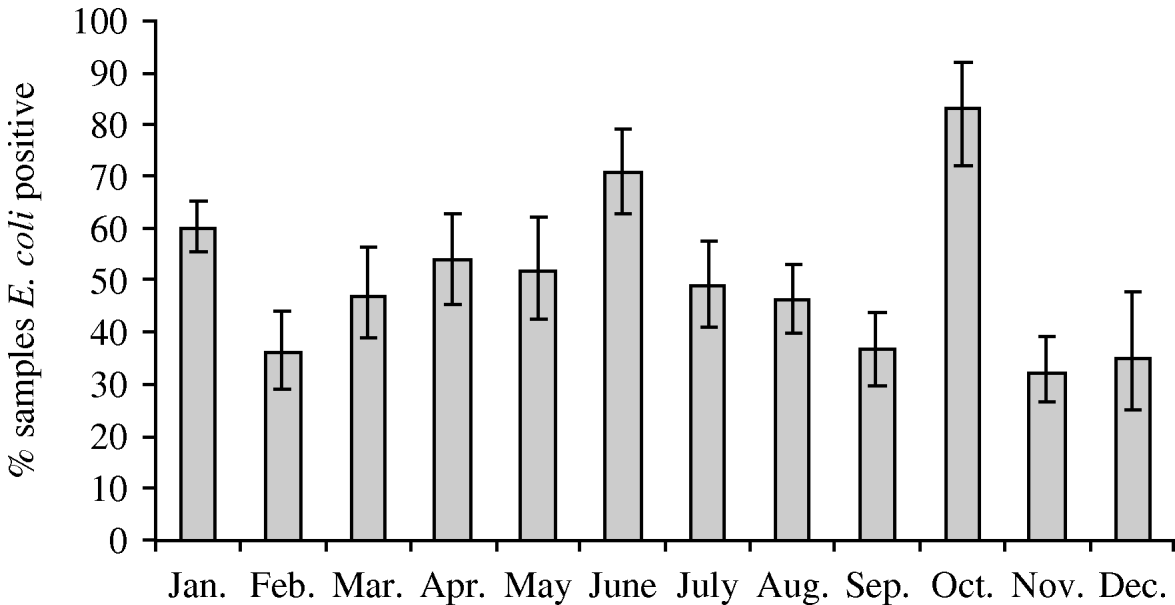
Fig. 1. Monthly prevalence estimates of E. coli isolated from wild bird faecal samples. Ninety-five percent confidence intervals are shown.
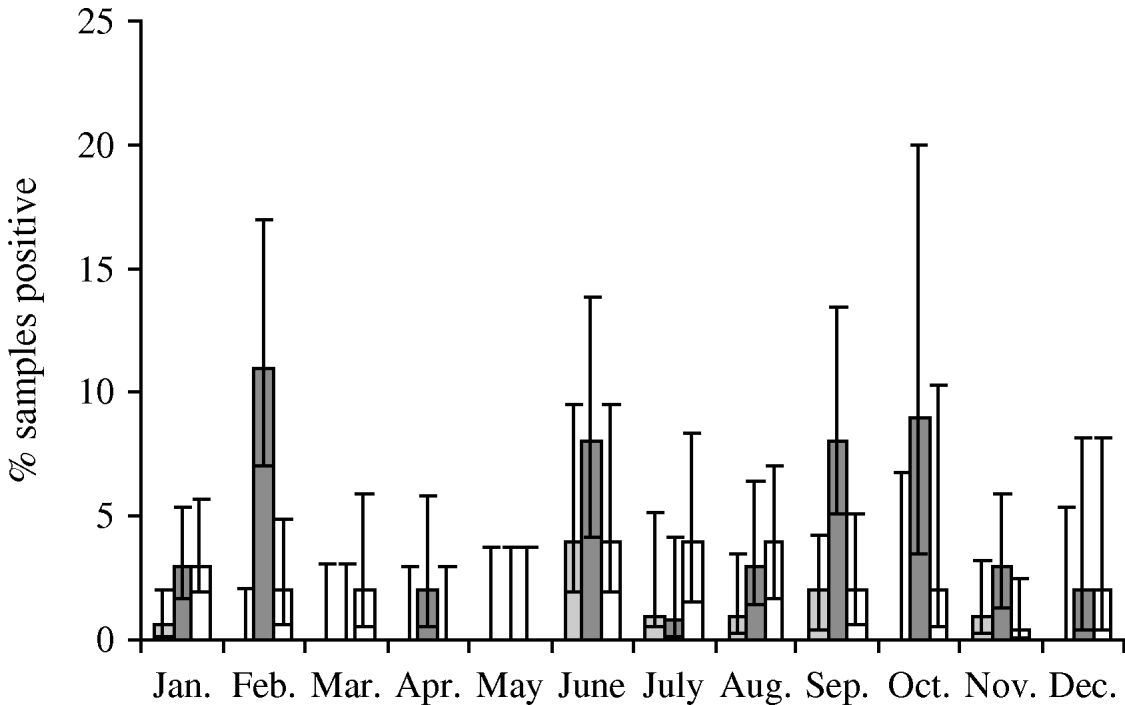
Fig. 2. Monthly prevalence estimates of E. coli isolated from wild bird faecal samples possessing virulence genes stx1 (![]() ), stx2 (
), stx2 (![]() ) and eae (□). Ninety-five percent confidence intervals are shown.
) and eae (□). Ninety-five percent confidence intervals are shown.
Multivariable analysis
Risk factors for the occurrence of E. coli containing stx1, stx2 and eae genes
The location at which birds were sampled was included in all multivariable logistic regression models as a random effect to account for the potential clustering effect. However, the proportion of the variation attributable to sampling location was negligible in all cases.
Table 1 shows the final multivariable logistic regression model of risk factors associated with the occurrence of E. coli in which stx1 genes were detected. The occurrence of stx1 genes was associated with the year during which samples were collected: the odds of isolating E. coli possessing stx1 genes from faecal samples were more than four times greater in 2005 than in 2004, and stx1 genes were not detected in E. coli isolated during 2006. There were increased odds that E. coli positive for stx1 genes were isolated from faecal samples collected during summer (June–August inclusive) compared to winter (December–February inclusive). Faecal samples from which Salmonella was isolated were more likely to yield E. coli containing stx1 genes. In addition, there was a positive association between the occurrence of stx1 genes and eae genes in E. coli isolated from faecal samples. Goodness-of-fit statistics provided evidence for a good model fit (Hosmer–Lemeshow χ2=1·62, P=0·81; area under the ROC curve=0·78).
Table 1. Factors associated with the isolation of E. coli possessing the virulence gene stx1 from wild bird faecal samples as determined by multivariable logistic regression analysis (n=1196)

OR, Odds ratio; CI, confidence interval.
The final multivariable logistic regression model of risk factors associated with the occurrence of E. coli possessing stx2 genes is shown in Table 2. The odds of isolating E. coli containing stx2 genes from faecal samples were significantly lower during spring (March–May inclusive) and significantly higher during autumn (September–November inclusive) compared to winter. Compared to samples collected from Anseriformes (ducks, geese and swans), samples collected from Passeriformes (passerines) had reduced odds of being positive for E. coli containing stx2 genes; however, stx2 genes were more likely to be detected in E. coli isolated from Falconiformes (falcons) and Charadriiformes (wading birds). E. coli containing stx2 genes were more likely to be isolated from faecal samples from which Salmonella was also isolated. The Hosmer–Lemeshow diagnostic test statistic was 7·55 (P=0·18) and the area under the ROC curve was 0·81 indicating that the model was a good fit.
Table 2. Factors associated with the isolation of E. coli possessing the virulence gene stx2 from wild bird faecal samples as determined by multivariable logistic regression analysis (n=1748)
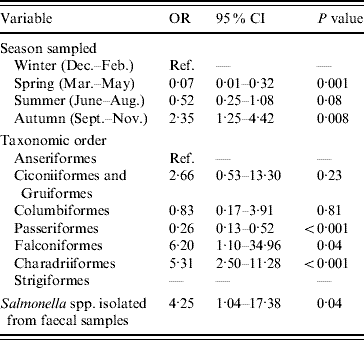
OR, Odds ratio; CI, confidence interval.
The final multivariable model of risk factors associated with the occurrence of E. coli in which eae genes were detected is shown in Table 3. Compared to samples collected during 2004, there were significantly lower odds of isolating E. coli with eae genes from samples collected during 2006. E. coli containing eae genes were more likely to be present in samples collected in autumn compared to samples collected during winter. Passeriformes had lower odds of carrying E. coli with eae genes when compared to Strigiformes (owls). Last, E. coli containing eae genes were more likely to be isolated from faecal samples from which E. coli containing stx1 genes were also detected. The Hosmer–Lemeshow diagnostic test statistic for the multivariable model was 7·84 (P=0·35) and the area under the ROC curve was 0·73 indicating that the model was a good fit.
Table 3. Factors associated with the isolation of E. coli possessing the virulence gene eae from wild bird faecal samples as determined by multivariable logistic regression analysis (n=1748)
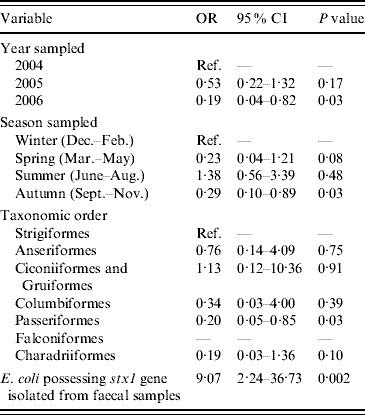
OR, Odds ratio; CI, confidence interval.
DISCUSSION
The occurrence of STEC has been reported in wild birds previously, but studies have tended to concentrate on detecting particular serotypes of E. coli known to be associated with disease in humans, e.g. O157:H7, or detecting virulence genes in a limited number of wild bird species. Consequently, little is known about the wider host range or prevalence of STEC virulence determinants in E. coli carried by wild bird populations. E. coli virulence genes, known to be associated with human disease, namely stx1, stx2 and eae were found in wild bird samples at a prevalence of 1·5% (95% CI 0·9–2·5), 7·9% (95% CI 6·4–9·8) and 4·9% (95% CI 3·7–6·4), respectively. This is consistent with previous studies where stx genes have been detected in wild birds at a prevalence ranging from 1·6% to 12·5% and eae genes have been reported in wild birds at a prevalence ranging from 4·5% to 40·0% [Reference Pedersen16–Reference Kobayashi, Pohjanvirta and Pelkonen18, Reference Nielsen27–Reference Schmidt29]. Domestic animals, in particular cattle, are considered to be the main reservoir of human STEC infections [Reference Chapman3]. The results of our study are comparable with those of a study carried out over the same time period and in the same locality as the present study, which detected stx1-, stx2- and eae-positive E. coli in cattle faecal samples at prevalences of 3·0%, 1·8% and 5·4%, respectively (A. Lahuerta-Marin, personal communication).
The risk factors associated with the occurrence of stx1 and stx2 genes in E. coli isolated from wild birds differed, suggesting that these virulence genes, which are transmitted between bacteria on highly mobile bacteriophages, may have differing ecologies and may not occur at random. The proportion of E. coli isolates in which stx1 genes were detected varied significantly during the 3 years of the study and the occurrence of eae genes declined significantly each year. However, it is difficult to determine if these observed differences reflect trends, and if so what might drive such trends. Similar year-to-year variability has been reported in the occurrence of STEC serogroup O157 in cattle and it was concluded that studies carried out over a longer time period would be required to determine if the observed variation was a genuine trend [Reference Gunn30]. Year on year variation in environmental factors and the dynamics of wild bird populations, E. coli and bacteriophages may all be involved. The interplay between such factors can be complex.
The occurrence of each virulence gene was associated with a different season in the final multivariable models. The odds of E. coli containing stx1 genes being isolated from wild birds were almost seven times greater during summer (June–August) than during winter (December–February), whereas there were reduced odds of isolating E. coli containing stx2 genes during spring (March–May) and increased odds during autumn (September–November). The association of stx-positive E. coli with particular seasons is difficult to explain. It could be that clonal STEC strains may have been circulating among wild bird populations during those seasons. It could also be that other unidentified risk factors, for which season is acting as a proxy measure, influence the shedding patterns of E. coli containing stx and eae genes in wild bird populations. Again, these factors (e.g. diet) might affect the dynamics of E. coli infection and shedding and/or the dynamics of stx-encoding bacteriophages.
There are very few studies of the occurrence of STEC in wild birds that report any seasonal trend. A study of the occurrence of variant stx2 gene stx2f in feral pigeons in Italy reported no association between the occurrence of E. coli possessing the stx2f gene and season [Reference Morabito17]. However, another study reported a higher frequency of eae genes in E. coli isolated from rock pigeons in Colorado, USA, during summer and autumn compared with winter [Reference Pedersen16]. This contrasts with the findings of the present study where the odds of detecting eae were significantly less during autumn than in winter. As already mentioned, there are likely to be complex interactions between hosts (different birds and different environments) and microorganisms (strains of E. coli and bacteriophages carrying stx genes), which give rise to the observed seasonal patterns that are likely to vary over time.
Multivariable logistic regression analysis revealed a significant association between the occurrence of E. coli containing stx2 genes and the taxonomic order of the bird. No such association was found for the occurrence of stx1-positive E. coli. Stx2-positive E. coli were detected in faecal samples collected from 30 species of wild bird. However, they were most strongly associated with samples collected from Charadriiformes (wading birds) and Falconiformes (falcons). Most wading birds commonly feed in estuaries and it is possible that estuarine habitats are a source of stx2-positive E. coli, which birds may acquire through ingestion: 84% of birds sampled in an estuarine habitat in this study were Charadriiformes. It was not possible to enter the habitat in which birds were sampled into the final multivariable model as there was a correlation between the habitat in which birds were sampled and the taxonomic order. However, univariable analysis showed that stx2 genes were most commonly detected in E. coli isolated from birds sampled in estuarine habitats (data not presented).
The increased frequency of stx2-positive E. coli from birds sampled in estuarine habitats could be a result of such bacteria being present in rivers that discharge into estuaries. Previous studies have reported the occurrence of stx2 genes in both coliform populations [Reference Garcia-Aljaro31] and as free bacteriophages [Reference Muniesa, Serra-Moreno and Jofre32] in aquatic environmental samples, including rivers. Earlier studies showed that waterways can be contaminated with E. coli containing virulence genes from slaughterhouse wastewater [Reference Loukiadis33] and by direct faecal contamination by livestock [Reference Ihekweazu34].
Under certain conditions, such as a lack of nutrients and ultraviolet light irradiation, temperate lambdoid bacteriophages present within E. coli can be induced from the lysogenic cycle into the lytic cycle causing the release of infectious bacteriophages [Reference Muniesa7]. It is possible that the conditions required to induce stx2-bacteriophages into the lytic cycle are provided by estuarine habitats resulting in the release of free bacteriophages into the environment. The results of several studies indicate that stx2-bacteriophages have a diverse host range of E. coli strains; therefore, estuarine habitats may be a source of stx2-gene-containing E. coli and infectious bacteriophages, which birds acquire through feeding.
The association between stx2-positive E. coli with Falconiformes may also be explained by diet. Wild bird species that comprised the Falconiform group in this study included the common buzzard (Bueto bueto), common kestrel (Falco tinnunculus), peregrine (Falco peregrinus) and Eurasian sparrowhawk (Accipiter nisus). All of these birds are predatory and feed on small mammals, carrion, passerine birds, wading birds and/or pigeons. There are few studies that report STEC in wildlife species; however, stx-gene positive E. coli have been reported previously in rodents [Reference Nielsen27], rabbits [Reference Scaife35], passerines [Reference Nielsen27], pigeons [Reference Morabito17, Reference Kobayashi, Pohjanvirta and Pelkonen18, Reference Schmidt29] and the present study found a strong association between stx2-positive E. coli and wading birds. It is likely that Falconiformes acquire stx2-gene-carrying E. coli via the oro-faecal route from their prey species.
Likewise the association between the occurrence of E. coli containing eae genes and Strigiformes (owls) may be diet related. The final logistic regression model showed that eae gene-positive E. coli were less likely to be isolated from Passeriformes (eae gene detected in 2% of samples) than from Strigiformes (eae gene detected in 19% of samples). The Strigiform group in this study was comprised mainly of barn owls (Tyto alba), which have a diet primarily of small rodents. A recent study that investigated the role of wildlife, including mammals and birds, in the transmission of STEC to livestock found a 5% prevalence of eae-positive E. coli in wild rodents (A. Lahuerta-Marin, personal communication). Barn owls generally swallow their prey whole providing a mechanism for the acquisition of eae-positive E. coli harboured in the intestines of their rodent prey.
Multivariable logistic regression revealed that stx1- and stx2-positive E. coli were more likely to occur in faecal samples that also contained Salmonella enterica. Salmonellae are yet to be associated with phage-related disease and it is therefore difficult to explain this association. It is could be that other unidentified risk factors, for which the occurrence of S. enterica is acting as a proxy, influence the occurrence of stx-positive E. coli in wild bird populations. For example, intestinal conditions that are favourable for the presence of E. coli carrying stx genes may also be favourable for the presence of S. enterica. Alternatively, wild birds from which both S. enterica and stx-positive E. coli were isolated may have acquired both bacteria from one source, e.g. livestock. Asymptomatic infection with both S. enterica and STEC is known to occur in some species of livestock, in particular ruminants [Reference Chapman3, Reference Edrington36].
The occurrence of the stx1 gene in E. coli isolated during the present study was highly associated with the occurrence of the eae gene. This association is difficult to explain as stx1 genes are encoded on bacteriophages whereas the eae gene is located on the LEE pathogenicity island on the bacterial chromosome. However, in a study investigating the prevalence of the eae gene in STEC strains from dairy cattle, eae-positive isolates were also more likely to be positive for stx1 [Reference Sandhu37].
In summary, we have shown that E. coli containing genes that encode Shiga toxin (stx1 and stx2) and intimin production (eae), which are important virulence determinants in E. coli associated with human disease, can be detected in a range of wild bird species in Great Britain. The stx2 gene was the most commonly detected virulence gene, and this has been shown by others [Reference Paton and Paton12, Reference Boerlin38] to be more strongly associated with severe human disease than stx1 genes. Wild bird populations represent a potential reservoir of virulence genes that are known to be transmitted horizontally between E. coli strains on highly mobile temperate lambdoid bacteriophages. This, coupled with the ability to act as long-distance vectors of STEC, means that wild birds have the potential to influence the spread and evolution of STEC. Logistic regression analysis identified several unique risk factors for the occurrence of each virulence gene, which may suggest these virulence genes do not occur at random and the mobile genetic elements that encode virulence genes have individual and differing ecologies.
ACKNOWLEDGMENTS
The authors thank the Merseyside Ringing Group for collection of field samples, RSPCA Stapeley Grange Wildlife Centre, the Garden Bird Health Initiative (British Veterinary Association Animal Welfare Foundation, CJ Wildbird Foods Ltd, Cranswick Pet Products, Gardman Ltd, RSPB, The Birdcare Standards Association, Universities Federation for Animal Welfare) and members of the public who submitted dead birds for post-mortem examination. This work was funded through the Defra VTRI programme. Laura Hughes has a Scholarship from the Faculty of Veterinary Science, University of Liverpool.
DECLARATION OF INTEREST
None.







