INTRODUCTION
Yersiniosis is a zoonotic gastrointestinal infection reported in humans worldwide. In the European Union it is the third most frequently reported zoonosis after campylobacteriosis and salmonellosis. Most reported infections are caused by Yersinia enterocolitica with only a few being due to Yersinia pseudotuberculosis [1]. The symptoms of yersiniosis are age-dependent and in children aged <5 years the most common symptom is non-specific gastroenteritis. For other age groups acute mesenteric lymphadenitis, septicaemia and sequelae such as arthritis, erythema nodosum and Reiter's syndrome have also been reported [Reference Bottone2]. The most frequent Y. enterocolitica bioserotypes pathogenic to humans are 1B/O:8, 2/O:5, 27, 2/O:9, 3/O:3 and 4/O:3, with the latter predominating in cases reported worldwide. Pathogenicity in Y. enterocolitica is linked to the presence of genes situated on both the chromosome and the plasmid [Reference Fredriksson-Ahomaa, Stolle and Korkeala3]. Since the presence of the plasmid is unstable, PCR primers and probes have frequently been directed towards a chromosomally located gene, for example the ail (attachment invasion locus) gene, as a more reliable target for detection [Reference Jourdan, Johnson and Wesley4–Reference Thisted Lambertz6]. Recently, two TaqMan PCR methods, targeting different locations on the ail gene in Y. enterocolitica and Y. pseudotuberculosis, respectively, were developed [Reference Thisted Lambertz6, Reference Thisted Lambertz, Nilsson and Hallanvuo7]. These methods were used for detection of the pathogens in this study.
The source of yersiniosis is considered to be contaminated food, especially pork and pork products [Reference Ostroff8, Reference Tauxe9]. Pigs are the only food animals that regularly harbour the pathogen [Reference Bhaduri, Wesley and Bush10–Reference Lindblad12] and are considered the main reservoir [Reference Hurvell, Glatthard and Thal13, Reference Wauters14]. To develop control measures and lower the incidence of Y. enterocolitica or maintain Yersinia-free herds, better knowledge is needed of possible sources of contamination at the farm level. Wild rodents may be one such source, since these pests often have free access to pig houses. Previous studies have shown that Y. enterocolitica is common in wild rodents, but isolation of the most widespread human pathogenic bioserotype, 4/O:3, is rare [Reference Aldova, Cerny and Chmela15–Reference Kapperud17].
Y. pseudotuberculosis is a foodborne pathogen causing repeated outbreaks in certain countries in the Northern Hemisphere, e.g. Finland, Canada, Russia and Japan [Reference Hallanvuo18]. Although wild birds, rodents and pigs appear to be major reservoirs, this pathogen seems to generally circulate in the environment between water, soil and wild animals [Reference Hallanvuo18, Reference Fukushima and Gomyoda19]. In Finland there are almost annually occurring outbreaks of Y. pseudotuberculosis [Reference Hallanvuo18], some of which have been traced to carrots or iceberg lettuce, possibly contaminated by wild animals [Reference Jalava20–Reference Nuorti22]. Y. pseudotuberculosis has also been isolated from pigs and pork products and in some of these cases rodents have been suspected of carrying and spreading the infection [Reference Laukkanen23].
Due to inefficient methods, pathogenic strains of Y. enterocolitica are often difficult to isolate, especially from environmental samples. The underlying reason is that in these samples the pathogen is present in low numbers together with a high amount of background flora, and sufficiently selective culture media are not available [Reference Fredriksson-Ahomaa and Korkeala24]. Instead, PCR methods can be utilized and currently the use of probe-based real-time PCR has improved the specificity of the analysis. However, in certain studies, it is necessary to isolate the bacterium; e.g. in outbreak investigations or when tracing a source of contamination it is crucial to compare the genotypes of the isolates involved. At present the gold standard for genotyping pathogenic Y. enterocolitica and Y. pseudotuberculosis is pulsed-field gel electrophoresis (PFGE) [Reference Jalava20, Reference Laukkanen23, Reference Asplund, Johansson and Siitonen25–Reference Thisted Lambertz and Danielsson-Tham27]. Random amplified polymorphic DNA (RAPD) is a rapid genetic fingerprinting method that can be used in addition to PFGE. It has previously been tested on foodborne Y. enterocolitica spp. [Reference Blixt28].
The objectives of this study were to investigate the presence of pathogenic Y. enterocolitica and Y. pseudotuberculosis in wild rodents caught at various locations in Sweden, including pig farms, using TaqMan PCR for detection and conventional culture for isolation of the two pathogens; and two fingerprinting methods to identify and compare the genotypes of the isolates recovered.
MATERIALS AND METHODS
Sampling procedures
Between December 2005 and December 2007, rodent traps were set at 28 locations in Sweden: on 16 pig farms, five chicken farms, and seven other non-farm-related locations. A total number of 207 rodents were caught at 20 of the 28 trapping locations, while none were caught at the remaining eight locations. Eight were discarded from the analysis due to technical problems. Both live and snap traps (traps that instantly kill the rodent) were set at points with signs of rodent activity, such as burrows or droppings. Number of traps used and number of days the traps were set at specific locations varied depending on the area and supply of rodents, i.e. between 10 and 50 traps and between 2 days and several weeks. Traps were checked every day. Most of the trapping locations (n=24) were in central/southeast Sweden (Mälardalen region) but four of the pig farms were located in southern and southwest Sweden (Småland/Halland region). The capture and euthanasia of rodents were approved by the Swedish Ethical Committee for Scientific Experiments (protocol C247/5).
Both rodents and pigs were examined for the presence of pathogenic Y. enterocolitica and Y. pseudotuberculosis. The occurrence in rodents was studied by examining colon tissue samples from 199 rodents and lymph nodes from 128 of the same rodents. Nine colon samples were lost during processing. The sampled rodents originated from seven pig farms (n=110), five chicken farms (n=55) and six other locations (n=25). Information regarding the capture locations and rodent species caught is listed in Table 1. At necropsy, performed on the day of capture, about 1–1·5 cm of proximal colon and superficial cervical lymph nodes from all rodents were aseptically removed and collected in separate 1·5-ml microcentrifuge tubes. Samples were immediately transported to the laboratory and either processed on the same day or stored at −80°C until analysed. Histological examination of intestines, kidney, liver, lungs and spleen was performed at the Department of Pathology and Wildlife Diseases, National Veterinary Institute (Uppsala, Sweden).
Table 1. Locations where rodents were sampled and proportion of rodents that tested positive by TaqMan PCR for pathogenic Y. enterocolitica
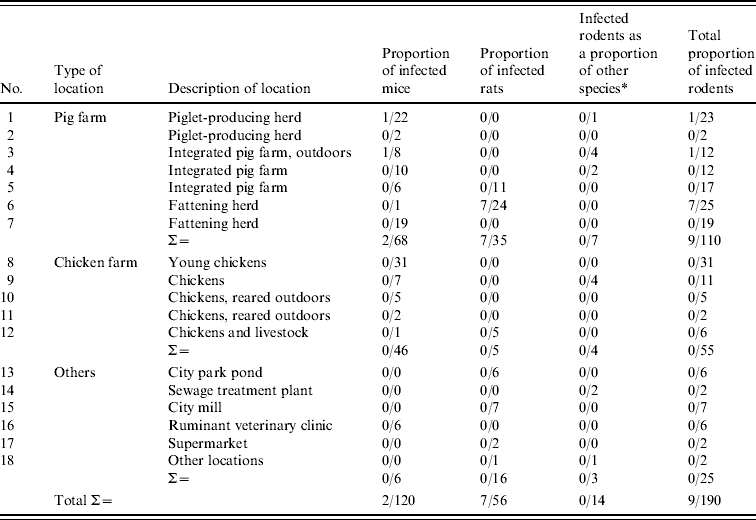
* Thitreen yellow-necked mice, Apodemus flavicollis and one water vole, Microus agrestis (location 18).
The occurrence of pathogenic Y. enterocolitica in pigs was studied for three of the seven pig farms where rodents were caught, i.e. locations 3, 5 and 6 (Table 1); fattening pigs at locations 3 and 6, and growers at location 5. On each of the three farms, 20 individual live pigs were sampled by Amies culture swabs (Copan Innovation, Italy). The swabs were rubbed over the rectal mucosa covering an area of about 2 cm×2 cm. Swabs were stored in Amies agar gel medium with charcoal (Copan Innovation) at 8°C until cultivation.
Sample preparation and culture
The rodent colon tissue samples and the superficial cervical lymph nodes were treated with 150–250 μl of 0·9% NaCl solution and the tissues were thoroughly mashed with a Pasteur pipette to achieve a homogeneous mixture. Samples were then vortexed and centrifuged (50 g) for 10 min. Sub-portions (10 μl) of the supernatant from each sample were spread onto Cefsulodin Irgasan Novobiocin (CIN) agar plates (Oxoid, CM 653 and SR 109) and the remaining volume was used for DNA extraction (see below). The pig swabs were streaked directly onto CIN agar plates. All plates were incubated at 30°C for 21±3 h. If no typical colonies had appeared after 24 h, the CIN agar plates were incubated another 21±3 h. Small red ‘bull's-eye’ colonies were considered presumptive pathogenic Y. enterocolitica. Up to four colonies per plate were subcultured and transferred to individual tubes containing brain heart infusion (BHI) broth (Oxoid, CM0225) mixed with 17% glycerol and stored at −80°C until characterization.
Positive control strains
In the TaqMan PCR methods, strain SLV-408 (CCUG 45643) of Y. enterocolitica 4/O:3 and strain TAVA 81 of Y. pseudotuberculosis were used as reference and control strains. Strain SLV-408 was also used as positive control when performing the bioserotyping and PFGE. Sterile distilled water was used as a negative control in the PCR.
DNA extraction and TaqMan PCR
DNA was extracted from the tissue samples on the remaining supernatants (as described above) with the DNeasy Blood and Tissue kit (Qiagen GmbH, Germany). Moreover, following subculture on BHI agar of the isolated colonies recovered from the rodents and pigs, DNA was prepared by transfer of a loop of the bacteria to 200 μl sterile distilled water with 20 μl of 0·8 m NaOH solution added. The tubes were incubated at 75°C for ~10 min and 48 μl of equal volumes of 0·8 m HCl and 0·1 m Tris (pH 8·3), were added [Reference Thisted Lambertz6]. In some cases and when preparing DNA for RAPD analysis, again the DNeasy Blood and Tissue kit was used according to the manufacturer's protocol for Gram-negative bacteria. For the detection of pathogenic Y. enterocolitica and Y. pseudotuberculosis, two TaqMan probe-based PCR methods targeting different sites of the ail gene were applied as described previously [Reference Thisted Lambertz6, Reference Thisted Lambertz, Nilsson and Hallanvuo7]. A positive amplification control was included in the analysis [Reference Thisted Lambertz6]. In cases of PCR inhibition, which occurred more frequently in the lymph node samples than the colon samples, samples were diluted 1:10 and re-tested. Monitored by the amplification control, this was sufficient to eliminate the effect of any inhibitory substances present.
Presumptive colonies isolated from the rodent and pig samples were initially examined by PCR for identification of pathogenic Y. enterocolitica. PCR-negative colonies were further analysed by TaqMan PCR for identification of Y. pseudotuberculosis.
Phenotypic analysis
Isolates testing positive in TaqMan PCR for pathogenic Y. enterocolitica were biotyped according to a reduced variant of the scheme by Wauters et al. [Reference Wauters, Kandolo and Janssens29], which included tests for lipase, salicin, esculin, xylose, trehalose, Voges–Proskauer and pyrazinamidase, performed as described previously [Reference Thisted Lambertz and Danielsson-Tham27]. To reveal presence or absence of the virulence plasmid, Congo Red-brain heart infusion agarose plates (CR-BHO) were used [Reference Bhaduri30]. The biotyped isolates were serotyped by a slide agglutination test with the commercial antisera O:3 and O:9 (Reagensia AB, Sweden).
Sequencing
PCR products obtained from three Y. enterocolitica isolates (68, 104, 200; see Table 2), three Y. enterocolitica TaqMan-positive colon samples from rodents, and one Y. pseudotuberculosis TaqMan-positive rodent colon sample, were sequenced at Uppsala Genome Centre (Rudbeck Laboratory, Sweden). The PCR products were purified with QIA quick PCR Purification kit (Qiagen GmbH). Sequencing was performed in both directions.
Table 2. Results of phenotypic and genotypic characterization of Y. enterocolitica 4/O:3 strains isolated from rodents and pigs in this study
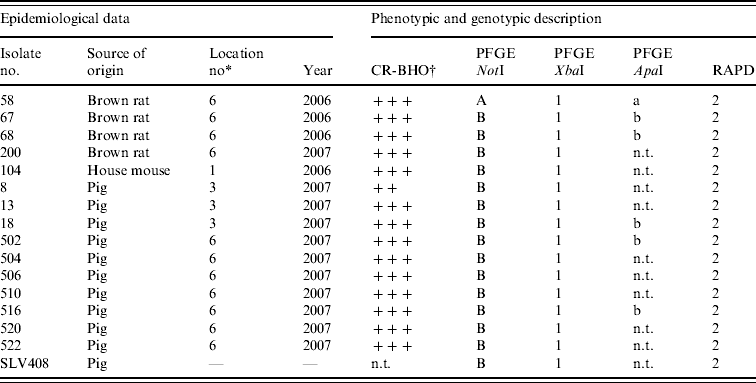
n.t., Not tested; CR-BHO, Congo Red-brain heart infusion agarose plates.
* Described in Table 1.
† CR-BHO results; numbers of colonies indicated as ++, 10–50; +++, >150 [Reference Thisted Lambertz and Danielsson-Tham27].
PFGE
The Y. enterocolitica 4/O:3 strains isolated from the rodent (n=5) and pig samples (n=10) were compared by PFGE. SLV-408 (CCUG 45643) was included as a control strain. The PFGE Standardized Laboratory Protocol for Molecular Subtyping of Escherichia coli O:157:H7, non-typhoidal Salmonella serotypes and Shigella sonnei [31, Reference Ribot32] was used according to the protocol with the following modifications: 300 μl instead of 2 ml cell suspension buffer, 100 μl instead of 200 μl restriction enzyme mixture containing 6·5 μg BSA and 1 μl restriction enzyme instead of 5 μl. To compensate for the smaller amount of enzyme, incubation time was extended to 4 h instead of 1·5–2 h as suggested in the protocol. Restriction enzymes NotI and XbaI (New England Biolabs, USA) were chosen and additional typing was performed for some of the isolates with ApaI. Salmonella serotype Braenderup H9812 was used as a standard [Reference Hunter33]. Electrophoresis was performed in 0·5× TBE buffer with 1–25 s switching time for 22 h at 14°C with a CHEF-DR III instrument (Bio-Rad Laboratories, USA). Gels (Agarose NA, 17-0554-02; Amersham Biosciences, UK) were stained with GelRed [Biotium (www.biotium.com)] and visualized with Quantity One software (Bio-Rad). PFGE patterns were analysed by visual examination of banding differences, and also analysed with GelCompare II software [Applied Maths (www.applied-maths.com)].
RAPD
All strains isolated from rodents and pigs were also analysed by RAPD. Ready-To-Go RAPD Analysis Beads (Pharmacia Biotech, USA) were used. The protocol provided by the manufacturer was used with the primers provided (5′-GGTGCGGGAA-3′ and 5′-GTTTCGCTCC-3′). PCR products were loaded on a 1% agarose gel stained with ethidium bromide and visualized digitally with GelDoc 2000 (Bio-Rad) and Quantity One software (Bio-Rad). Isolates differing in size and numbers of bands were assigned to different RAPD types.
RESULTS
Pathogenic Y. enterocolitica in rodents and pigs
The TaqMan PCR screening of the 190 rodent samples for presence of pathogenic Y. enterocolitica revealed that nine (5%) of 190 colon samples analysed, from rodents caught at locations 1, 3 and 6, were PCR-positive for pathogenic Y. enterocolitica (Table 1). All lymph node samples were negative. All PCR-positive samples originated from rodents caught on pig farms located at distances ranging 20–65 km from each other.
Conventional culture on the 190 rodent colon samples identified five colonies recovered from five individual rodents caught at locations 1 and 6 as Y. enterocolitica 4/O:3 by bioserotyping (Table 2). The results were confirmed by PCR. Thus, of the nine rodent samples that initially tested positive by PCR, five Y. enterocolitica 4/O:3 strains were obtained. From the pig swabs, 11 presumptive colonies from 11 individual pigs were confirmed as pathogenic Y. enterocolitica by TaqMan PCR, 4/20 pigs from location 3, and 7/20 pigs from location 6. Bioserotyping identified 10 of these colonies as 4/O:3 (Table 2), while one showed inconsistent serotyping results. Further analysis indicated that this particular strain of 4/O:3 was contaminated with Citrobacter freundii, and it was excluded from further analysis. The CR-BHO agarose plate analyses indicated that all 15 rodent and pig isolates identified as 4/O:3 harboured the virulence plasmid. Histological examination revealed no certain signs of infection in any of the rodents.
Y. pseudotuberculosis in rodents and pigs
The TaqMan PCR analyses for detection of Y. pseudotuberculosis resulted in one positive sample of 190 rodent colon samples analysed, whereas all 128 lymph node samples analysed tested negative. The positive sample came from a house mouse caught on a pig farm (location 3). No isolate was obtained. At autopsy, this mouse showed hyperaemic mucous membranes of the caecum and, histologically, moderate to severe acute multifocal hepatitis and splenitis. None of the isolates obtained from pigs tested positive in TaqMan PCR.
Sequencing
When the amplified 163-bp PCR products were sequenced, almost complete base-pair sequences, i.e. 160, 159, 163 and 159 bp, were obtained, from the three rodent Y. enterocolitica 4/O:3 strains and from one of the colon tissue samples, respectively. A similarity search in GenBank (www.ncbi.nlm.nih.gov) showed that they were identical to the corresponding parts of the deposited ail genes of Y. enterocolitica AY004311 and AJ605740. For the remaining two colon samples, the 52-bp short sequence obtained from one was identical to corresponding parts of AY004311, whereas the other sequence was of too poor quality to be analysed. The 159-bp sequence from the colon sample that tested positive for Y. pseudotuberculosis was identical to the ail gene of Y. pseudotuberculosis (acc. nos: CP001048, CP000720, BX936398).
PFGE and RAPD analyses
Results from the genotyping obtained by PFGE and RAPD are summarized in Table 2. PFGE analysis of 15 strains isolated from rodents (n=5) and pigs (n=10) generated one profile when cleaved with restriction enzyme XbaI and two profiles A and B when cleaved with restriction enzyme NotI (Fig. 1). The use of ApaI, which was applied for a selection of six of the isolates, produced two pulsotypes, a and b. The two groups of pulsotypes obtained with ApaI corresponded to the two groups of pulsotypes obtained with NotI. Just one rodent isolate showed the pulsotypes A and a, while all pig isolates, four rodent isolates and the control strain showed the pulsotypes B and b. Analyses of the same six isolates by RAPD showed one type of banding pattern for each of the two primers used. Thus, the discriminatory power was the same as for XbaI.
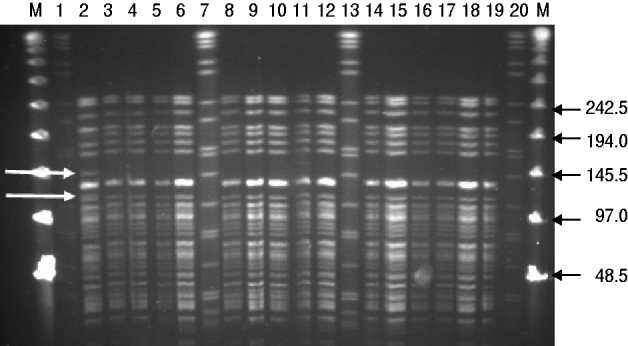
Fig. 1. PFGE (NotI) profiles for isolates of Y. enterocolitica 4/O:3 from rodents and pigs. M, Lambda marker. Lanes 1, 7, 13, 20, Standard Salmonella Braenderup H9812; lanes 2–5, rat isolates from location 6; lane 6, mouse isolate from location 1; lanes 8–10, pig isolates from location 3; lanes 11, 12, 14–18, pig isolates from location 6; lane 19, control strain SLV408. White arrows to the left indicate differences between pulsotypes A and B. Black arrows to the right indicate size of lambda marker bands.
DISCUSSION
In this study rodents were collected at different locations in Sweden (including pig farms) and their potential role as carriers of pathogenic strains of Y. enterocolitica was investigated. Y. enterocolitica bioserotype 4/O:3, which is the most common bioserotype reported in human yersiniosis throughout the world, was detected in about 5% at all locations, and on pig farms, in 8% of the rodents. The pathogen was detected in both mice and rats, but only in those caught on pig farms. The proportion of positive rats (20%) on pig farms is comparable to other studies that showed a prevalence in black rats on pig farms of between 14% and 17% [Reference Aldova, Cerny and Chmela15, Reference Pokorna and Aldova34]. However, in earlier studies, isolates of Y. enterocolitica from rodents were only serotyped and not biotyped and it is therefore uncertain whether those isolates were human pathogenic yersiniae [Reference Aldova, Cerny and Chmela15, Reference Pokorna and Aldova34, Reference Iinuma35]. In an early study where O:3 isolates recovered from field vole (Microtus agrestis) were biotyped, the biotypes obtained differed from those recognized as being human pathogens [Reference Kapperud17]. Over the years, knowledge of the pathogenic determinants of the pathogen has increased and new techniques have been introduced, so that detection of the pathogenic bioserotypes is now both rapid and specific. Simultaneously, the workload in performing biochemical tests for biotyping has been reduced. We found it useful to first apply a TaqMan PCR method for screening the rodent samples, to obtain an early indication of presence/absence of the pathogen in a sample. TaqMan PCR and biochemical reactions were then applied on the presumptive colonies appearing on CIN agar [Reference Wauters, Kandolo and Janssens29], and then isolates were serotyped and the virulence plasmid-associated phenotypes of the colonies were determined with CR-BHO agarose [Reference Bhaduri30]. Besides reducing the time involved, this strategy made the confirmation steps more efficient in identification of the human pathogenic bioserotypes of the bacterium.
Y. enterocolitica 4/O:3 has not been reported in free-living rodents, but it has been isolated from rats living in proximity to pigs [Reference Kaneko36]. The results in the present study support these findings in that all 4/O:3-positive rodents identified were caught on pig farms and that rodents collected at other locations were found to be negative. Pathogens were predominately found in rats in this study. However, a few mice caught on pig farms also carried the pathogen. To our knowledge this is the first reported isolation of Y. enterocolitica 4/O:3 from a house mouse. The fact that only rodents caught near pigs tested positive indicates that rather than being reservoirs, rodents are more likely to act as carriers of bacteria they contract from infected pigs and their environment. However, since the number of trapped animals on other locations was generally lower than on pig farms, this assumption should be made with some caution. On one farm, rat faeces were visible both inside and outside pig pens. Rats have been video-recorded feeding from the floor in pig pens [Reference Akande37], showing that faecal–oral transmission of the bacteria is likely to occur between pigs and rodents. The bacterium was isolated from rats caught in two consecutive years on the same farm, showing that colonization of rats is not an exceptional event. Generally, the farms with positive rodents were farms where rodents seemed to be abundant, based on information from farmers and visual signs of their presence. All farms in this study, like most Swedish farms, applied pest control by the use of rodenticides, but control of the rodent population was insufficient in some cases. Based on the data derived from this study, a high abundance of wild rodents in pig farms should always be regarded as a risk factor for maintaining pathogenic Y. enterocolitica infection in pigs. A recommendation to pig producers is to always emphasize pest control, including construction and maintenance of functional barriers.
PFGE revealed two pulsotypes among the 4/O:3 strains isolated from rodents and pigs in this study. One of the pulsotypes originated from a single strain isolated from one of the rats, while the DNA profiles of the remaining strains deriving from four rodent isolates were indistinguishable and similar to those derived from the pig isolates. In an attempt to improve the discriminatory power, in addition to using the two restriction enzymes NotI and XbaI, the restriction enzyme ApaI was applied as suggested by Fredriksson-Ahomaa et al. [Reference Fredriksson-Ahomaa, Autio and Korkeala26]. The use of RAPD with two sets of primers showing identical patterns confirmed the similarity among the isolates. However, no additional differentiation was reached. This is in agreement with previous studies where the usefulness of RAPD in differentiating between Yersinia strains was poor [Reference Blixt28]. In Japan Hayashidani et al. [Reference Hayashidani38] isolated the highly virulent bioserotype 1B/O:8 of Y. enterocolitica from rodents and pigs and revealed similar pulsotypes in the rodent and pig isolates, suggesting a common source of contamination. In contrast to Y. enterocolitica 4/O:3, bioserotype 1B/O:8 can be found in the environment and has repeatedly been isolated from free-living wild rodents of different species [Reference Iinuma35]. While rodents may be regarded as reservoirs for 1B/O:8 [Reference Iinuma35, Reference Hayashidani39], thereby also constituting a direct risk for public health, rodents carrying 4/O:3 strains appear more likely to be vectors for pathogen transmission between pigs within a pig herd, as indicated by the present study and others [Reference Pokorna and Aldova34, Reference Kaneko36].
Y. pseudotuberculosis was detected in only one of the rodent samples examined (1/190) and in none of the 60 pig samples, indicating a low prevalence of this pathogen in these animals in Sweden. Similarly, Y. pseudotuberculosis is only rarely reported as a source of human infection in Sweden and no human outbreaks have been reported. In contrast, recent studies have shown that in Finland, pigs most probably play a role as a reservoir of human Y. pseudotuberculosis infections [Reference Niskanen, Fredriksson-Ahomaa and Korkeala40] and that pest animals may be responsible for spreading the bacterium on Finnish pig farms [Reference Laukkanen23]. However, Y. pseudotuberculosis is difficult to detect by available detection methods and therefore can easily be overlooked. It often persists in low numbers and it is debatable whether the direct detection approach applied in the present study was sensitive enough to reveal the pathogen.
In conclusion, the results obtained in our study suggest that rodents, primarily the brown rat and to a lesser extent the house mouse, are possible vectors for transmission of Y. enterocolitica 4/O:3 on pig farms. Since there is no evidence of rodents acting as reservoirs of the infection, they should mainly be considered as posing a risk for maintaining and spreading the bacteria within a farm, especially between different batches of pigs in all-in/all-out systems.
ACKNOWLEDGEMENTS
The study was funded by grants from the Swedish Farmers' Foundation for Agricultural Research and the Swedish Research Council (Formas). Material supplies were kindly provided by National Food Administration, Sweden and Anticimex. We also thank Ricardo Feinstein, Department of Pathology, National Veterinary Institute, Sweden, for histological examination of samples, and Viveca Båverud, Department of Bacteriology, National Veterinary Institute, and Karl-Erik Johansson, Department of Biomedical Sciences and Veterinary Public Health, Swedish University of Agricultural Sciences, for valuable inputs on the manuscript.
DECLARATION OF INTEREST
None.





