Lung cancer is one of the most frequently diagnosed forms of cancer, as well as the leading cause of cancer-related death among males, whereas it is the fourth most commonly diagnosed cancer and the second leading cause of cancer-related death among females worldwide (Jemal et al., Reference Jemal, Bray, Center, Ferlay, Ward and Forman2011). Non-small cell lung cancer (NSCLC) might account for 85% of these cases, which mainly include squamous cell carcinoma (SCC), adenocarcinoma (AC), and large cell carcinoma (Zandberga et al., Reference Zandberga, Kozirovskis, Ābols, Andrējeva, Purkalne and Linē2013). Although the etiology and the mechanisms of lung cancer remain uncertain, it seems likely that the development of lung cancer is a multi-factor and multi-step process. Tobacco smoking greatly increases one's risk of developing lung cancer, in addition to environmental exposure, such as ionizing radiation and ultraviolet rays, which can cause bulky adducts, cross-links, and deoxyribonucleic acid (DNA) damage (Sakoda et al., Reference Sakoda, Loomis, Doherty, Julianto, Barnett, Neuhouser and Chen2012). However, research has also indicated that individual genetic variation might increase one's susceptibility to lung cancer.
Excision repair cross-complementing group 2 (ERCC2), also called Xerodermapigmentosum group D (XPD), is located on chromosome 19q13.3 and comprises 23 exons and spans across 54,000 base pairs; this group includes important DNA damage repair genes, which play an important role in nucleotide excision repair (NER) pathways (Weber et al., Reference Weber, Salazar, Stewart and Thompson1990). DNA damage, caused by exogenous carcinogens or endogenous carcinogens, plays an important role in the carcinogenesis process (Dong et al., Reference Dong, Zhuang and Ma2013). However, NER pathways repair a broad variety of DNA damage, including oxidative damage, cross-links, and bulky adducts, such as polycyclic aromatic hydrocarbon and other aromatic compounds (Sreeja et al., Reference Sreeja, Syamala, Syamala, Hariharan, Raveendran, Vijayalekshmi and Ankathil2008). Researchers have identified the high frequency Lys751Gln (rs13181) of ERCC2 mutations in protein regions. The most important change occurred between the ERCC2 encoded protein and its helicase activator, p44 protein, inside the transcription factor II H (TFIIH) complex, where A to C substitution in exon 23 might result in an amino acid alteration from lysine (Lys) to glycine (Gln). The A→C variation might be associated with adduced repair capacity, which could facilitate cancer development (Benhamou et al., Reference Benhamou and Sarasin2002).
Researchers report that Gln variant alleles are associated with a lower defect repair capacity (DRC) of UV-induced DNA damage, as opposed to homozygous, wild-type alleles (Benhamou et al., Reference Benhamou and Sarasin2002). ERCC2 Lys751Gln polymorphism might lead to a defect in NER, deficient DRC, and an increased susceptibility to cancer. Previous studies have suggested that ERCC2 Lys751Gln polymorphism is associated with various cancers, such as esophageal cancer, cutaneous melanoma, and hepatocellular carcinoma (Ding et al., Reference Ding, Ma, He and Zhang2012; Dong et al., Reference Dong, Zhuang and Ma2013; Zhang et al., Reference Zhang and Mou2013). Shen et al. (Reference Shen, Berndt, Rothman, Demarini, Mumford, He and Lan2005) have shown that ERCC2 Lys751Gln is associated with a decreased risk of lung cancer, whereas Liang et al. (Reference Liang, Xing, Miao, Tan, Yu, Lu and Lin2003) have revealed that a 751Gln/Gln genotype may lead to an increased risk for lung cancer. Most studies, including recent meta-analysis (Manuguerra et al., Reference Manuguerra, Saletta, Karagas, Berwick, Veglia, Vineis and Matullo2006; Zhan et al., Reference Zhan, Wang, Wei, Wang, Qian, Yu and Song2010), have assessed the potential relationship of ERCC2 Lys751Gln polymorphism in the development of lung cancer. However, the results remain inconclusive and inconsistent, possibly due to a small sample size that is not adequate for investigating the relationship between them. We conducted a comprehensive, updated meta-analysis in order to assess the association between ERCC2 Lys751Gln polymorphism and lung cancer.
Materials and Methods
Search Strategy and Selection Criteria
We searched through PubMed and Embase for all eligible studies published prior to April 20, 2013, for ERCC2 Lys751Gln polymorphism and lung cancer risk, using the following terms: ‘Excision repair cross complementing group 2’, ‘ERCC2’, ‘Xerodermapigmentosum group D’, ‘XPD’, ‘Lys751Gln’, ‘rs13181’, ‘polymorphism’, and ‘lung cancer’. Our meta-analysis did not include any limitations on language and encompassed all human subjects. Eligibility criteria included: (1) using case-control designs, (2) estimating the association of the ERCC2 Lys751Gln polymorphism and lung cancer, (3) calculating the results with an odds ratio (OR) of 95% confidence interval (95% CI) in each study, and (4) providing sufficient genotype data for estimations. We excluded all case-only studies or reviews.
Data Extraction
According to the above criteria, two investigators (X. Tan and L. Xian) carefully extracted literature searches and data from all eligible studies, while a third party (M. W. Chen) made independent discrepancy decisions. The following characteristics were extracted from the included studies: the author/s’ name/s, country of origin, year of publication, ethnicity, source of controls, genotyping method, and the number of cases and controls, respectively.
Statistical Analysis
We estimated the strength of the association between the ERCC2 Lys751Gln polymorphism and lung cancer risk using the pooled OR and 95% CI in the following genetic models: allele contrast (Gln allele vs. Lys allele), homozygote comparison (CC vs. AA), heterozygote comparison (CA vs. AA), recessive model (CC vs. CA+AA), and dominant model (CC+CA vs. AA).
Using a chi-square-based Q statistic test and I 2 statistic test, we checked the between-study heterogeneity (Higgins et al., Reference Higgins, Thompson, Deeks and Altman2003). No heterogeneity existed, according to calculations using the p value > .1 and I 2 < 25%, whereas I 2 > 50% indicated that heterogeneity was moderate. We used the fixed-effects model to conduct Mantel-Haenszel's method (Hu et al., Reference Hu, Zhang, Wang, Li, Li and Zhang2013), whereas we found that the random effects model with the DerSimonian and Laird method must be performed when I 2 > 50% or p value < .1 (Lau et al., Reference Lau, Ioannidis and Schmid1997). We estimated the Hardy-Weinberg equilibrium (HWE) using a chi-square test in the genotype distribution of ERCC2 Lys751Gln among the controls. The equation p > .05 revealed that the distribution of genotypes among the controls agreed with HWE (Bosco et al., Reference Bosco, Castro and Briones2012). We then estimated the potential publication bias using Begg's funnel plot and Egger's linear regression test. No publication bias existed for a p value > .05 (Peters et al., Reference Peters, Sutton, Jones, Abrams and Rushton2006).
We conducted an assessment of our methodology using the Newcastle-Ottawa Scale (NOS) in our meta-analysis. The NOS included eight items, divided into three dimensions: selection, comparability, and study type exposure (case-control studies) or outcome (cohort studies). A star system may be used for semi-quantitative evaluation of the studies quality. We allowed the comparability group to reach two stars, whereas we awarded each other item a maximum of one star in the highest quality studies. We used NOS ranges from zero to nine stars to estimate the values, though standard criteria have not been established (Stang et al., Reference Stang2010). We considered each article gained seven or more than seven stars as a high quality article in our research.
In order to evaluate the effects of covariance, we performed subgroup analyses, determined by ethnic subgroup (Asian and Caucasian), control source (population-based and hospital-based), and pathologic types (SCC and AC). All analyses were carried out using Stata software, version 11.1 (Stata Corporation, USA). All of the p values were two-sided, wherein less than .05 was considered statistically significant.
Results
Characteristics of Studies
We identified a total of 29 relevant studies via our PubMed and Embase research and by scanning the abstracts (the publication selection process is shown in Figure 1). However, we excluded one of the studies after checking the full-text articles due to a lack of data (Park et al., Reference Park, Lee, Jeon, Park, Bae, Lee and Jung2002). We ultimately included 28 studies in our meta-analysis. One study contained two ethnicities, divided into two individual studies (Chang et al., Reference Chang, Wrensch, Hansen, Sison, Aldrich, Quesenberry and Wiencke2008; David-Beabes et al., Reference David-Beabes, Lunn and London2001). Our research encompassed 28 full studies (Chang et al., Reference Chang, Wrensch, Hansen, Sison, Aldrich, Quesenberry and Wiencke2008; Chen et al., Reference Chen, Tang, Xue, Xu, Ma, Hsu and Cho2002; David-Beabes et al., Reference David-Beabes, Lunn and London2001; De Ruyck et al., Reference De Ruyck, Szaumkessel, De Rudder, Dehoorne, Vral, Claes and Thierens2007; Harms et al., Reference Harms, Salama, Sierra-Torres, Cajas-Salazar and Au2004; Hou et al., Reference Hou, Fält, Angelini, Yang, Nyberg, Lambert and Hemminki2002; Hu et al., Reference Hu, Xu, Shao, Yuan, Wang, Wang and Shen2006; Kiyohara et al., Reference Kiyohara, Horiuchi, Takayama and Nakanishi2012; Liang et al., Reference Liang, Xing, Miao, Tan, Yu, Lu and Lin2003; López-Cima et al., Reference López-Cima, González-Arriaga, García-Castro, Pascual, Marrón, Puente and Tardón2007; Matullo et al., Reference Matullo, Dunning, Guarrera, Baynes, Polidoro, Garte and Vineis2006; Misra et al., Reference Misra, Ratnasinghe, Tangrea, Virtamo, Andersen, Barrett, Taylor and Albanes2003; Osawa et al., Reference Osawa, Miyaishi, Uchino, Osawa, Inoue, Nakarai and Takahashi2010; Popanda et al., Reference Popanda, Schattenberg, Phong, Butkiewicz, Risch, Edler and Schmezer2004; Qian et al., Reference Qian, Zhang, Zhang, Zhou, Yu and Chen2011; Raaschou-Nielsen et al., Reference Raaschou-Nielsen, Sørensen, Overvad, Tjønneland and Vogel2008; Sakoda et al., Reference Sakoda, Loomis, Doherty, Julianto, Barnett, Neuhouser and Chen2012; Shen et al., Reference Shen, Berndt, Rothman, Demarini, Mumford, He and Lan2005; Spitz et al., Reference Spitz, Wu, Wang, Wang, Shete, Amos and Wei2001; Sreeja et al., Reference Sreeja, Syamala, Syamala, Hariharan, Raveendran, Vijayalekshmi and Ankathil2008; Vogel et al., Reference Vogel, Laros, Jacobsen, Thomsen, Bak, Olsen and Raaschou-Nielsen2004; Xing et al., Reference Xing, Tan, Wei and Lin2002; Yin et al., Reference Yin, Ma, Cui, Li, He and Zhou2006a; Yin et al., Reference Yin, Vogel, Ma, Guo, Wang and Qi2006b; Yin et al., Reference Yin, Su, Li, Li, Ma, He and Zhou2009; Zhou et al., Reference Zhou, Liu, Miller, Thurston, Xu, Wain and Christiani2002, Reference Zhou, Wan, Gao, Ding and Jun2012; Zienolddiny et al., Reference Zienolddiny, Campa, Lind, Ryberg, Skaug, Stangeland and Haugen2006), including 30 individual studies, with a total of 23,370 subjects (10,242 cases and 13,128 controls; Table 1). In the subgroup analysis, there were 14 studies on Caucasians and 13 studies on Asians (Table 1). Two studies, Chang et al. (Reference Chang, Wrensch, Hansen, Sison, Aldrich, Quesenberry and Wiencke2008) and David-Beabes et al. (Reference David-Beabes, Lunn and London2001), focused on African-Americans and Latino-Americans, respectively. Within the subgroup analysis, 18 studies included a population-based group and another 12 studies involved a hospital-based group (Table 1). Only some studies (Osawa et al., Reference Osawa, Miyaishi, Uchino, Osawa, Inoue, Nakarai and Takahashi2010; Popanda et al., Reference Popanda, Schattenberg, Phong, Butkiewicz, Risch, Edler and Schmezer2004; Xing et al., Reference Xing, Tan, Wei and Lin2002; Yin et al., Reference Yin, Ma, Cui, Li, He and Zhou2006b, Reference Yin, Su, Li, Li, Ma, He and Zhou2009) provided raw data concerning genotype distribution. Finally, we investigated the relationship between Lys751Gln polymorphism and pathologic types of lung cancer.
TABLE 1 Studies and Data Included in the Meta-Analysis
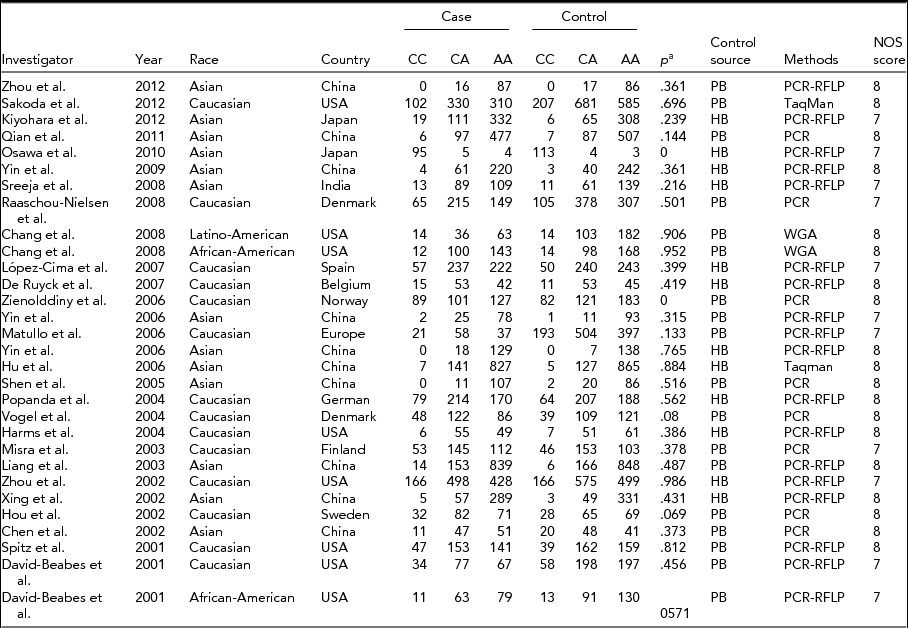
a p for Hardy–Weinberg equilibrium in control group; HB = hospital-based; PB = population-based; PCR = polymerase chain reaction; PCR-RFLP = polymerase chain reaction restriction fragment length polymorphism; WGA = whole genome amplification.
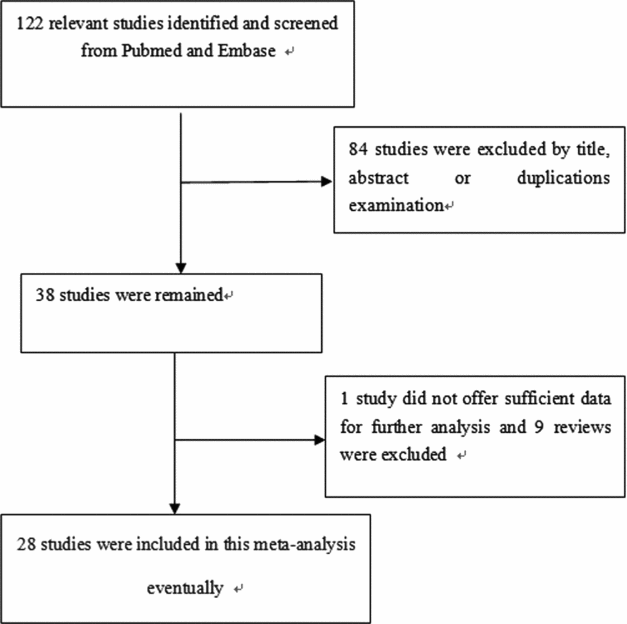
FIGURE 1 The publication selection process of this meta-analysis.
Meta-Analysis Results
The primary results from our meta-analysis on the relationship between ERCC2 Lys751Gln polymorphism and lung cancer are shown in Table 2. We considered using two studies (Osawa et al., Reference Osawa, Miyaishi, Uchino, Osawa, Inoue, Nakarai and Takahashi2010; Zienolddiny et al., Reference Zienolddiny, Campa, Lind, Ryberg, Skaug, Stangeland and Haugen2006) from the controls that disagreed with HWE, but they were excluded in the sensitivity analysis (Minelli et al., Reference Minelli, Thompson, Abrams, Thakkinstian and Attia2008). Therefore, our pooled analysis indicates that a carried Gln allele might be associated with an increased risk of lung cancer (Gln allele vs. Lys allele: OR = 1.160, 95% CI = 1.081–1.245, p = .000; Figure 2). Similarly, we find the same results under any of the contrast models (CC vs. AA: OR = 1.252, 95% CI = 1.130–1.388, p = .000; CA vs. AA: OR = 1.152, 95% CI = 1.060–1.252, p = .001; CC+CA vs. AA: OR = 1.186, 95% CI = 1.089–1.292, p = .000; CC vs. CA+AA: OR = 1.196, 95% CI = 1.087–1.316, p = .000; Table 2).
TABLE 2 The Results of the Meta-Analysis Lys751Gln Relation With Lung Cancer
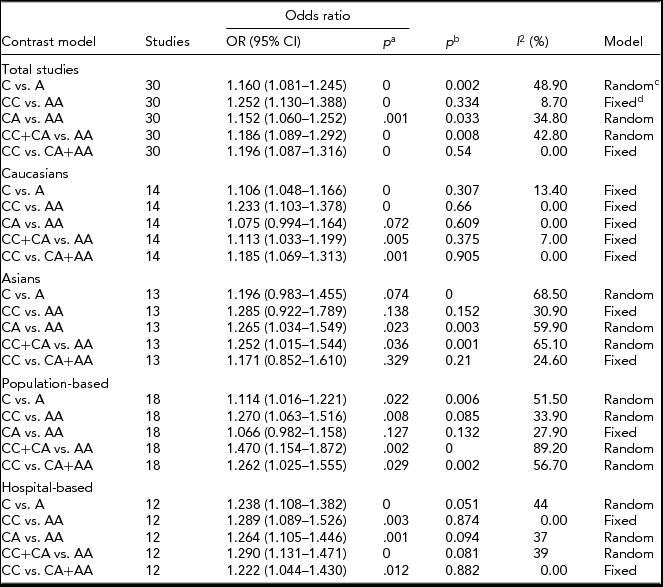
aThe pooled p value; b p value for heterogeneity test; crandom-effect model; dfixed-effect model.

FIGURE 2 Forest plots for the ERCC2 Lys751Gln polymorphism and risk of lung cancer in overall studies using the fixed-effect dominant model (C vs. A).
In the subgroup analysis based on Caucasians and Asians, there is a significant relationship between ERCC2 Lys751Gln polymorphism and susceptibility to lung cancer according to four contrast models among Caucasians (C vs. A: OR = 1.106, 95% CI = 1.048–1.166, p = .000; CC vs. AA: OR = 1.233, 95% CI = 1.103–1.378, p = .000; CC+CA vs. AA: OR = 1.113, 95% CI = 1.033–1.199, P = .005; CC vs. CA+AA: OR = 1.185, 95% CI = 1.069–1.313, p = .001; Table 2, Figure 3). Similarly, a significant association exists among Asians in two genetic models (CA vs. AA: OR = 1.265, 95% CI = 1.034–1.549, p = .023; CC+CA vs. AA: OR = 1.252, 95% CI = 1.015–1.544, p = .036; Table 2, Figure 4).
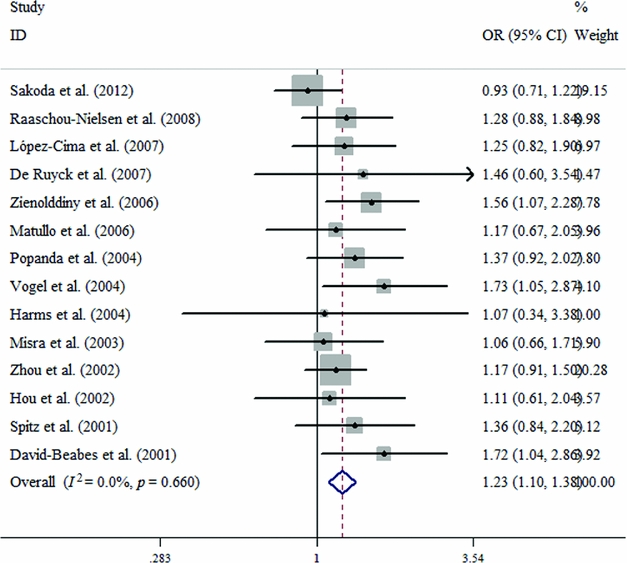
FIGURE 3 Forest plots for the ERCC2 Lys751Gln polymorphism and risk of lung cancer among Caucasians using the fixed-effect dominant model (CC vs. AA).
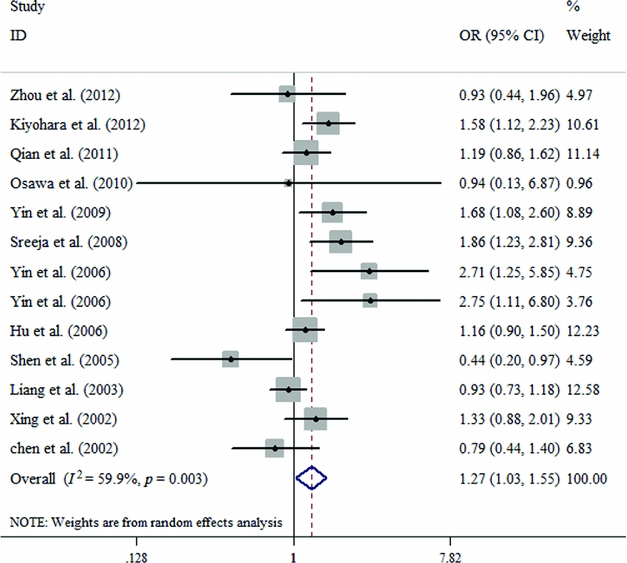
FIGURE 4 Forest plots for the ERCC2 Lys751Gln polymorphism and risk of lung cancer among Asians using the random-effect dominant model (CA vs. AA).
When we stratified our analysis by source of control, we found a statistically elevated risk in the hospital-based and population-based groups (Table 2). In our pathologic types analysis, we also found a significantly increased risk in the AC in three genetic models (C vs. A: OR = 1.295, 95% CI = 1.075–1.560, p = .006; CA vs. AA: OR = 1.366, 95% CI = 1.065–1.753, p = .014; CC+CA vs. AA: OR = 1.396, 95% CI = 1.099–1.773, p = .006), while in the SCC we also obtained the same results under any genetic models (C vs. A: OR = 1.720, 95% CI = 1.194–2.478, p = .004; CA vs. AA: OR = 1.780, 95% CI = 1.170–2.708, p = .007; CC+CA vs. AA: OR = 1.864, 95% CI = 1.242–2.799, p = .003).
Test for Heterogeneity, Sensitive Analysis and Test for Publication Bias
We could not detect any obvious heterogeneity in our overall pooled analyses; however, some genetic models in the Asian and population-based subgroups did display significant heterogeneity. We performed a sensitivity analysis by omitting every study once; every time, the heterogeneity did not decrease significantly.
Begg's funnel plot and the Egger's test were used to assess the potential publication bias. No evidence of publication bias were detected in the overall pooled studies (C vs. A: Begg's test, p = .354, Egger's test, p = .191; CC vs. AA: Begg's test, p = .646, Egger's test, p = .222; CA vs. AA: Begg's test, p = .269, Egger's test, p = .067; dominant model: Begg's test, p = .432, Egger's test, p = .145; recessive model: Begg's test, p = .416, Egger's test, p = .335; Figure 5).
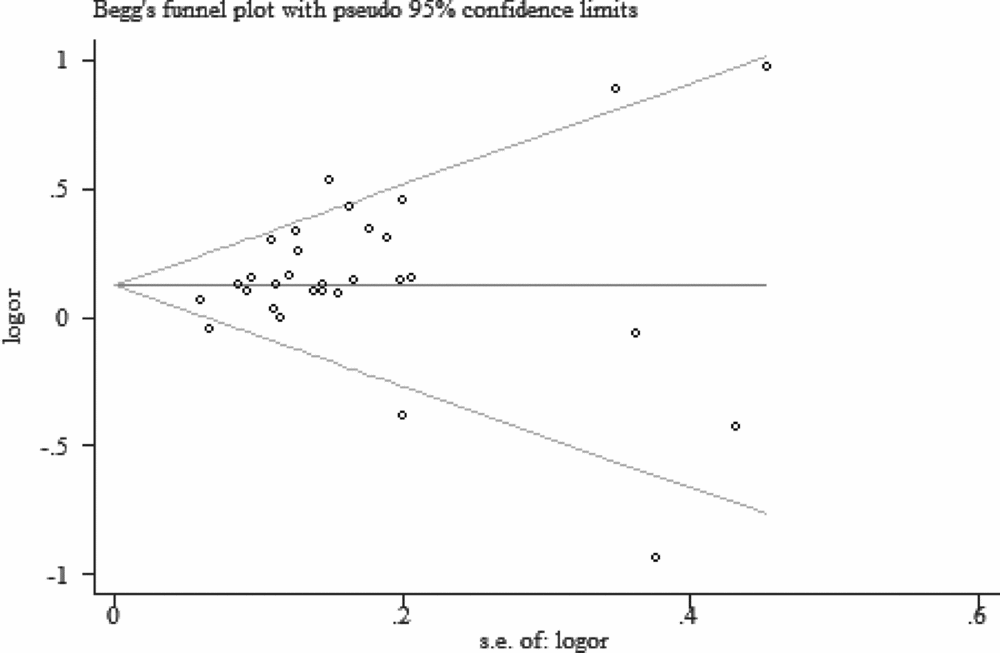
FIGURE 5 Begg's funnel plot of allele comparison for assessing the publication bias (C vs. A).
Discussion
Lung cancer is the leading cause of death via malignancy, and the most common among tobacco-induced cancers. Cigarette smoke contains a multitude of carcinogens and genotoxicants, including polycyclic aromatic hydrocarbons (PAH), such as benzo[a]pyrene (B[a]P) which could be bioactivated in vivo into benzo[a] pyrenedihydrodiol epoxide (BPDE), thus leading to irreversible DNA damage through oxidation or covalent binding (Wei et al., Reference Wei, Cheng, Hong and Spitz1996). BPDE-induced DNA adducts, UV-induced photoproducts, bulky mono-adducts, cross-links, and oxidative damage are all mostly repaired by the NER pathway (Spitz et al., Reference Spitz, Wu, Wang, Wang, Shete, Amos and Wei2001; Tang et al., Reference Tang, Pierce, Doisy, Nazimiec and MacLeod1992). Hemminki et al. (Reference Hemminki, Xu, Angelini, Snellman, Jansen, Lambert and Hou2001) revealed that Gln/Gln carriers, when compared to the wild-type Lys/Lys carriers, were 50% less efficient when repairing ultraviolet-specific cyclobutane pyrimidine dimmers. ERCC2 gene variations may restrain its protein products from interacting with p44, a subunit of TFIIH, thereby reducing its helicase activity, which leads to a NER defect (Taylor et al., Reference Taylor, Broughton, Botta, Stefanini, Sarasin, Jaspers and Lehmann1997). The ERCC2 gene plays an important role in NER pathway and basal transcription, and acts as an adenosine triphosphate (ATP)-dependent helicase in the multi-subunit transcription repair factor of the TFIIH complex (Spitz et al., Reference Spitz, Wu, Wang, Wang, Shete, Amos and Wei2001). Qiao et al. (Reference Qiao, Spitz, Shen, Guo, Shete, Hedayati and Wei2002) demonstrated that the DNA repair capacity (or DRC) among the subjects who were homozygous for the wild-type genotypes ERCC2 Lys/Lys, was significantly higher than those with variant genotypes Gln/Gln. Research suggests there is a five-fold variation in DRC within the general population, and decreased DRC may increase the risk of lung cancer (Hu et al., Reference Hu, Xu, Shao, Yuan, Wang, Wang and Shen2006).
Many case-control studies have investigated the correlation between ERCC2 Lys751G1n polymorphism and lung cancer. For example, Yin et al. (Reference Yin, Su, Li, Li, Ma, He and Zhou2009) have shown that the ERCC2 751AC/CC genotypes, as compared to those carrying the AA genotype, are associated with an increased risk for developing lung adenocarcinoma. David-Beabes et al. (Reference David-Beabes, Lunn and London2001) and Osawa et al. (Reference Osawa, Miyaishi, Uchino, Osawa, Inoue, Nakarai and Takahashi2010) did not find a statistically significant risk between the ERCC2 Lys751G1npolymorphism and lung cancer. Several meta-analyses recently investigated ERCC2 Lys751G1n polymorphism with lung cancer risk (Benhamou et al., Reference Benhamou and Sarasin2005; Hu et al., Reference Hu, Wei, Wang and Shen2004; Kiyohara et al., Reference Kiyohara and Yoshimasu2007; Manuguerra et al., Reference Manuguerra, Saletta, Karagas, Berwick, Veglia, Vineis and Matullo2006; Vineis et al., Reference Vineis, Manuguerra, Kavvoura, Guarrera, Allione, Rosa and Matullo2009; Wang et al., Reference Wang, Chang, Hu, Sui, Han, Li and Zhao2008; Zhan et al., Reference Zhan, Wang, Wei, Wang, Qian, Yu and Song2010; Zhang et al., Reference Zhang, Gu, Zhang, Jia and Chang2010). The latest meta-analysis that included a large sample size suggested that the ERCC2 751Gln variant allele significantly increased the risk of lung cancer for both Caucasians and Latino-Americans (Feng et al., Reference Feng, Ni, Dong, Shen and Du2012). However, the results are still controversial and inconclusive.
To the best of our knowledge, this is the largest sample size meta-analysis that comprehensively assesses the correlation between ERCC2 Lys751Gln polymorphism and lung cancer to date. Our meta-analysis demonstrates that ERCC2 Lys751Gln polymorphism is not only associated with lung cancer among Caucasians, but it is also associated with an increased risk of lung cancer among Asians. However, some of the research does not accord with the latest meta-analysis (Feng et al., Reference Feng, Ni, Dong, Shen and Du2012). Two studies deviated from HWE in our meta-analysis, so we performed a sensitivity analysis. When we omitted each study, the results of the re-analysis are stable (data not shown). In addition, we did not find a publication bias when we performed the Begg's and Egger's tests, which made our meta-analysis conclusions more credible.
The subgroup analyses for Caucasians and Asians, in addition to previous meta-analyses, show that ERCC2 Lys751Gln polymorphism is associated with lung cancer among Caucasians. There was also a significant association among Caucasians in the Gln allele, compared to the Lys allele (OR = 1.106, CI = 1.048–1.166). Our meta-analysis also provides new evidence that suggests that ERCC2 Lys751Gln polymorphism may be associated with an increased lung cancer risk among Asians under two genetic models (CA vs. AA: OR = 1.265, 95% CI = 1.034–1.549, p = .023 and CC+CA vs. AA: OR = 1.252, 95% CI = 1.015–1.544, p = .036). Our study reveals that the heterozygote CA carriers may increase the risk of developing lung cancer among Asians. Some additional factors could affect the development of cancer, including the fact that various ethnicities have different genetic polymorphisms and genetic backgrounds. In addition, previous reports may have used a low sample size, so large-scale subjects would be needed to investigate the correlations. The subgroup analyses that are based on source controls reveal a significant association between ERCC2 Lys751Gln polymorphism and lung cancer in the population-based and hospital-based groups. The source of the hospital-based subgroup might be associated with other diseases, particularly the genotypes investigated in the controls, which might lead to some biases in our results. Our research shows the same results in two groups. So we carefully and cautiously interpreted our results. In the pathologic types’ analyses, we found a significant association between AC and SCC, though we note that SCC might be induced by B[a]P or other PAH (Deutsch-Wenzel et al., Reference Deutsch-Wenzel, Brune, Grimmer, Dettbarn and Misfeld1983). Further research should be done to investigate the link between Lys751Gln polymorphism, SCC, and AC.
Although we made considerable efforts to collect all available data in our comprehensive analysis, some limitations should be acknowledged. First, although we included 23,370 subjects in our research, the sample size was relatively small in the stratified analyses, particularly for pathologic types. More research needs to be done to establish the relationship. Second, although the source of the controls did not differ, the inclusion criteria did not have a uniform definition. Healthy controls who were recruited from the same geographical area acted as the reference group for some studies, whereas others studies selected hospital patients without organic lung cancer as the reference group. Furthermore, age, gender, smoking status, cancer type, and ethnicity were not consistent in all studied subjects. As stated above, these factors may be sources of heterogeneity. Finally, although Begg's funnel plot and the Egger's test did not indicate any publication bias in our research, the tests excluded unpublished articles and abstracts, which might lead to a publication bias. In spite of this, our research is far more comprehensive than any single study to date.
In conclusion, this meta-analysis suggested that ERCC2 Lys751Gln polymorphism most likely contributes to an increased susceptibility to lung cancer risk among Caucasians and Asians alike. In addition, gene-gene and gene-environmental interactions within lung cancer should be studied further, because the existing data is insufficient. Therefore, further well-designed case-control studies with a large sample size are necessary to continue to investigate the correlation between ERCC2 polymorphisms and lung cancer development.
Acknowledgment
This work was supported by the Guangxi scientific research and technology development project (No. 10124001A-47).









