1. Cyanobacterial symbioses in land plants—an overview
Chloroplasts emerged as a result of a cyanobacterial symbiosis. From a single endosymbiotic event about 1.5 bya (Bengtson et al., Reference Bengtson, Sallstedt, Belivanova and Whitehouse2017; Butterfield, Reference Butterfield2000; Eme et al., Reference Eme, Sharpe, Brown and Roger2014), almost the entire diversity of photosynthetic eukaryotes radiated (Archibald, Reference Archibald2015). The nature of that cyanobacterial plastid progenitor is still debated (de Vries & Archibald, Reference de Vries and Archibald2017; Ponce-Toledo et al., Reference Ponce-Toledo, Deschamps, López-García, Zivanovic, Benzerara and Moreira2017); while some analyses suggest that the ancestor of a recently discovered unicellular cyanobacterium was the plastid progenitor (Ponce-Toledo et al., Reference Ponce-Toledo, Deschamps, López-García, Zivanovic, Benzerara and Moreira2017) other studies suggest that this ancestral free-living, symbiotically competent cyanobacterium might have been a filamentous cyanobacterium—as these analyses recover affiliation to a section of extant filamentous cyanobacteria based on phylogenetic and sequence similarity data (Dagan et al., Reference Dagan, Roettger, Stucken, Landan, Koch, Major, Gould, Goremykin, Rippka, Tandeau de Marsac, Gugger, Lockhart, Allen, Brune, Maus, Pühler and Martin2013; Ochoa de Alda et al., Reference Ochoa de Alda, Esteban, Diago and Houmard2014). Up until today, this section of filamentous cyanobacteria brings about symbiotically competent strains that are the foundation of diverse cyanobacterial symbioses—from fungal to plant hosts—that we find on our planet (Rai et al., Reference Rai, Soderback and Bergman2000). In these interactions, the cyanobacteria provide fixed nitrogen to their hosts.
Cyanobacterial symbionts (cyanobionts) of plants are present throughout the plant kingdom, yet they have emerged multiple times independently (Bergman et al., Reference Bergman, Matveyev and Rasmussen1996; Rai et al., Reference Rai, Soderback and Bergman2000; Figure 1). As a result, the degree of intimacy and the solutions on how the symbionts are housed are as different as the plants that undergo these symbioses (Figure 1): cyanobacterial symbioses of bryophytes are found among representatives from all major lineages, that is, mosses, liverworts and hornworts (Adams et al., Reference Adams, Bergman, Nierzwicki-Bauer, Duggan, Rai and Schüßler2013; Adams & Duggan, Reference Adams and Duggan2008). Feathermosses form epiphytic associations with cyanobionts. These are not ‘accidental’ associations, but the epiphytes are recruited and they fix nitrogen for the mosses (Bay et al., Reference Bay, Nahar, Oubre, Whitehouse, Wardle, Zackrisson, Nilsson and Rasmussen2013; Stuart et al., Reference Stuart, Pederson, Weyman, Weber, Rassmussen and Dupont2020). In contrast, hornworts and some liverworts (e.g., Blasia) have special mucilage-filled cavities, auricles or canals (hornwort Leiosporoceros dussii) to host cyanobacteria (Adams et al., Reference Adams, Bergman, Nierzwicki-Bauer, Duggan, Rai and Schüßler2013; Adams & Duggan, Reference Adams and Duggan2008). Other liverworts have only epiphytic associations (Adams & Duggan, Reference Adams and Duggan2008); yet they are not as well studied as those from feathermosses (Bay et al., Reference Bay, Nahar, Oubre, Whitehouse, Wardle, Zackrisson, Nilsson and Rasmussen2013; Ininbergs et al., Reference Ininbergs, Bay, Rasmussen, Wardl and Nisson2011; Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). Cycads (gymnosperms) induce the formation of a special organ type, the coralloid roots, that are colonised intercellularly by cyanobionts via broken tissue or lenticels (Bell-Doyon et al., Reference Bell-Doyon, Laroche, Saltonstall and Villarreal Aguilar2020; Rai et al., Reference Rai, Soderback and Bergman2000; Reinke, Reference Reinke1872; Suárez-Moo et al., Reference Suárez-Moo, Vovides, Griffith, Barona-Gómez and Cibrián-Jaramillo2019). The angiosperm Gunnera attracts its cyanobionts through mucilage-filled channels into their stems, where they enter and become intracellular (Khamar et al., Reference Khamar, Breathwaite, Prasse, Fraley, Secor, Chibane, Elhai and Chiu2010; Nilsson et al., Reference Nilsson, Rasmussen and Bergman2006; Towata, Reference Towata1985).
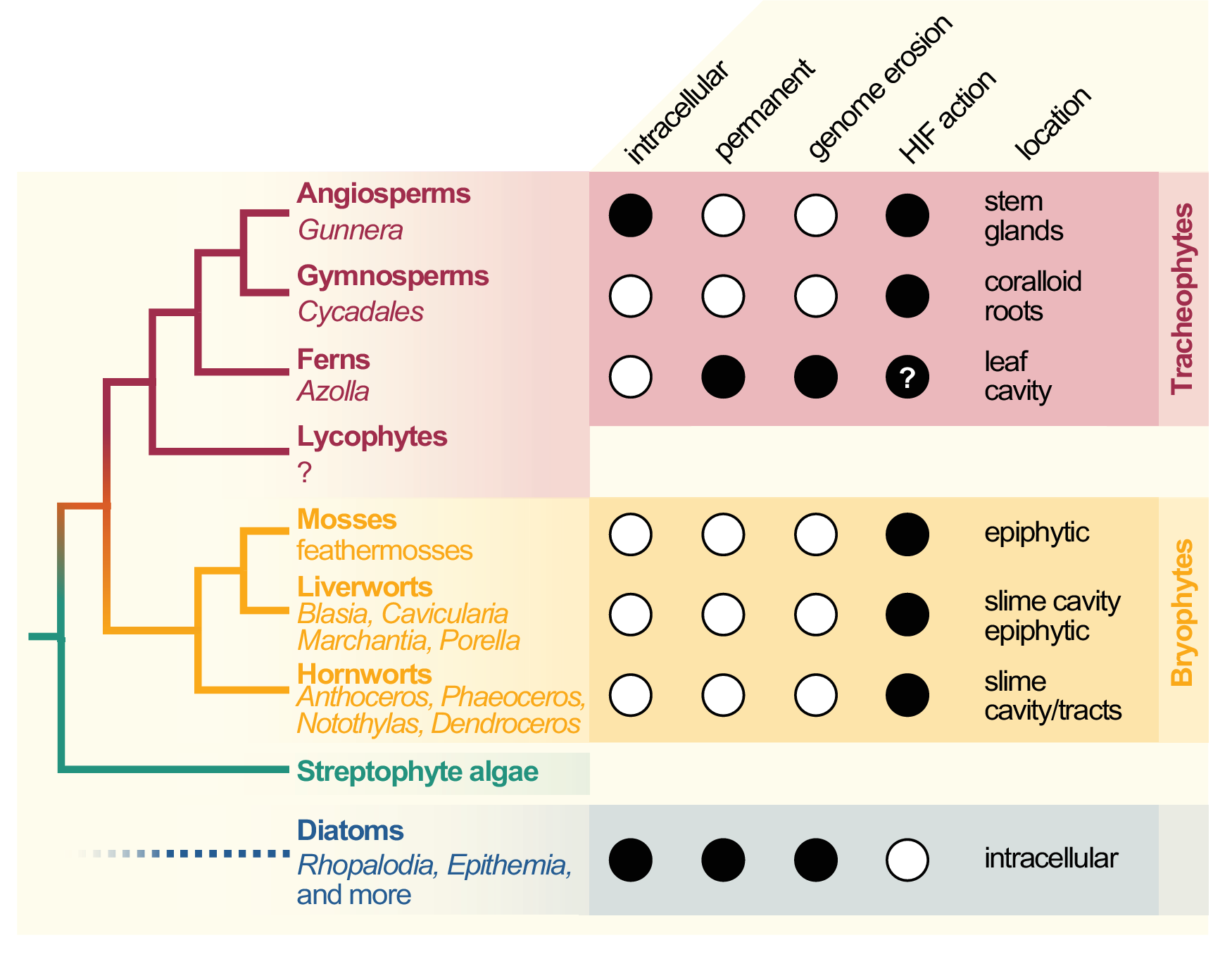
Figure 1. Cladogram of symbiotic interactions across land plants and diatoms. To the left, a simplified cladogram of the phylogenetic relationship among streptophytes (based on Puttick et al., Reference Puttick, Morris and Williams2018) is shown; diatoms (algae with a plastid of red algal origin) are very distantly related to streptophytes—the most recent common ancestor they share might be the last common ancestor of all eukaryotes (LECA). Among all major lineages of land plants except lycophytes (bold font), a few species that engage in symbiosis with cyanobacteria can be found (written in regular and italic font). To the right, filled dots denote key traits of the cyanobionts that dwell within those plants (and diatoms).
All aforementioned symbioses are facultative. In contrast to them stands the fern genus Azolla with their cyanobionts. Here, cyanobionts are transferred from one generation to the next (de Vries & de Vries, Reference de Vries and de Vries2018; Perkins & Peters, Reference Perkins and Peters1993; Zheng et al., Reference Zheng, Bergman, Chen, Zheng, Xiang and Rasmussen2009a). It is the only known permanent and obligate cyanobacterial symbiosis in the plant kingdom to date—that is apart from the chloroplast. Yet, in contrast to the chloroplast, the cyanobionts of Azolla are not endophytes, but live in special mucilage-filled cavities located in the dorsal leaf lobes (Figure 2).
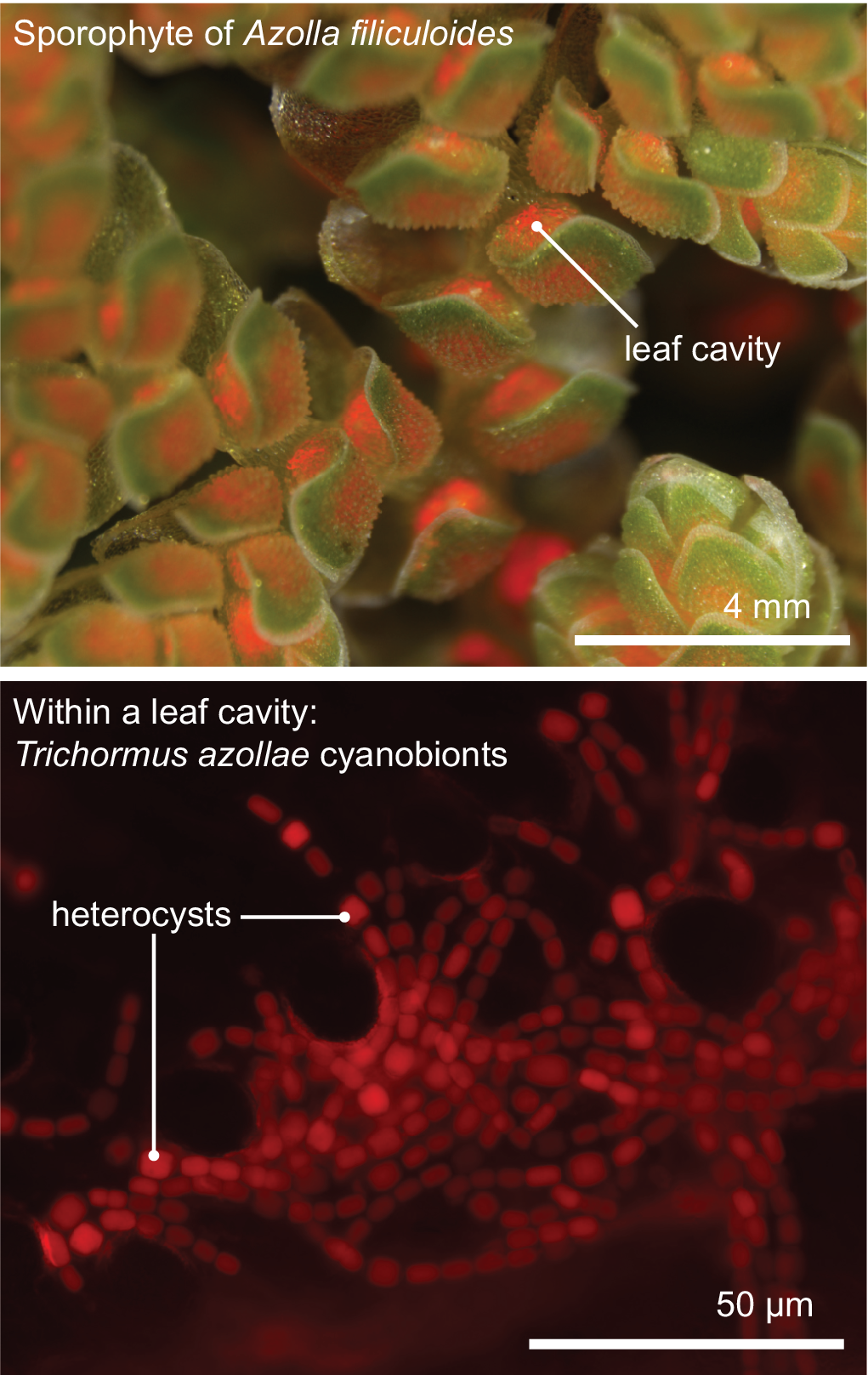
Figure 2. The Azolla–Trichormus symbiosis. At the top, the sporophyte body of Azolla filiculoides whose leaf cavities are packed with cyanobionts (red autofluorescence) is shown. At the bottom, a fluorescence micrograph of the filamentous cyanobionts (Trichormus azollae) within one leaf cavity are shown (red autofluorescence); note the heterocysts, which are larger than the vegetative cells.
Cyanobacterial symbioses do not occur in well-established model systems, although some of their hosts are currently emerging as such (Frangedakis et al., Reference Frangedakis, Shimamura, Villarreal, Li, Tomaselli, Waller, Sakakibara, Renzaglia and Szövényi2021; Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018). That said, the availability, the accessibility and the recent improvements to large-scale data have given the exploration of cyanobacterial symbioses a new boost (Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018; Reference Li, Nishiyama, Waller, Frangedakis, Keller, Li, Fernandez-Pozo, Barker, Bennett, Blázquez, Cheng, Cuming, de Vries, de Vries, Delaux, Diop, Harrison, Hauser, Hernández-García and Szövényi2020). Genomics, transcriptomics, proteomics and metagenomics provide new insights into the evolutionary biology of cyanobacterial symbioses. They establish the foundation to explore common recurrent patterns and identify the unique solutions that the evolutionary distant host plants have evolved to establish nitrogen-fixing symbioses with cyanobacteria. Comparative genomics on the cyanobacterial partners have identified patterns important in the biology of the bacteria, and those relevant for successful contributions to and gains from the symbiotic relationships. Here, we review the advances that the multiple different omics approaches have contributed to the understanding of cyanobacterial symbioses.
2. Recurrent molecular patterns in cyanobacterial symbioses: the cyanobionts
The scattered occurrences and diverse solutions in the establishment of the different plant-cyanobiont interactions may appear overwhelming. Yet, the questions that guide research on these symbioses are the same regardless of the symbiotic system. One of the key questions in plant-cyanobiont symbioses is what factors contribute to the initiation of the interaction. Here, much attention has been paid to the cyanobionts. Comparative genomics between symbiotically competent and not-competent cyanobacteria, as well as historically fingerprinting and nowadays metagenomic analyses, have been used to identify symbiotic communities and their features. Here, studies identified many recurring patterns.
2.1 Metagenomics, fingerprinting and cyanobacterial diversity
The question which cyanobacteria colonise the different hosts has led to several fingerprinting studies and isolation and Sanger sequencing of the cyanobacteria (e.g., Costa et al., Reference Costa, Paulsrud and Lindblad1999; Ininbergs et al., Reference Ininbergs, Bay, Rasmussen, Wardl and Nisson2011; Nilsson et al., Reference Nilsson, Bergman and Rasmussen2000; Zheng et al., Reference Zheng, Nilsson, Bergman and Rasmussen1999). Independent of the host, section IV/V cyanobacteria, foremost those of the order Nostocales, are consistently found in cyanobacterial symbioses (e.g., Liaimer et al., Reference Liaimer, Jensen and Dittmann2016; Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019; Pratte & Thiel, Reference Pratte and Thiel2021; Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010; Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). This suggests some inert symbiotic property that comes with the cyanobacteria of these sections. Specifically, most of the cyanobionts have been identified either as Nostoc or Anabaena (Adams et al., Reference Adams, Bergman, Nierzwicki-Bauer, Duggan, Rai and Schüßler2013; Rai et al., Reference Rai, Soderback and Bergman2000). The exception to the Nostoc/Anabaena symbionts is the cyanobiont of Azolla, which is as of late been called Trichormus azollae (see also discussions in Pereira & Vasconcelos, Reference Pereira and Vasconcelos2014). These symbiotic Nostocales are all filamentous, nitrogen-fixing and heterocyst-forming cyanobacteria. Some of them are able to colonise and associate in symbioses with various distinct hosts (Adams et al., Reference Adams, Bergman, Nierzwicki-Bauer, Duggan, Rai and Schüßler2013), suggesting that there may be limited host specificity.
Recent metagenomic data from hornworts uncovered that cyanobiont communities of the same host species from distinct locations can be quite different (Bouchard et al., Reference Bouchard, Peñaloza-Bojacá, Toupin, Guadalupe, Gudiño, Salazar Allen, Li and Villarreal Aguilar2020; Nelson et al., Reference Nelson, Hauser and Li2021). Location appears to play a major role in which (cyano)bacterial community is established in the symbiotic space. Even in the hornwort L. dussii, which has specialised canals that run through the entire lobe, geographic location of the host is the major determinant of the cyanobiont community, suggesting that the ability to colonise those canals is not uncommon for symbiotically capable cyanobacteria co-occurring with L. dussii (Bouchard et al., Reference Bouchard, Peñaloza-Bojacá, Toupin, Guadalupe, Gudiño, Salazar Allen, Li and Villarreal Aguilar2020). What cyanobiont will colonise the symbiotic space of hornworts is likely determined by the community present in the soil (Nelson et al., Reference Nelson, Hauser and Li2021). Despite that, the soil harbours certain species that never colonise the host, suggesting that while soil type dictates what species can generally become cyanobionts, the host has some measure of selectivity to allow only certain species entry into its symbiotic space (Nelson et al., Reference Nelson, Hauser and Li2021). This appears to be a common theme for the microbiome in tissues with close contact to soil. Other metabarcoding studies have highlighted that soil type is the major determined of the bacterial and eukaryotic root microbiota of plants (Lundberg et al., Reference Lundberg, Lebeis, Paredes, Yourstone, Gehring, Malfatti, Tremblay, Engelbrektson, Kunin, del Rio, Edgar, Eickhorst, Ley, Hugenholtz, Green Tringe and Dangl2012; Sapp et al., Reference Sapp, Ploch, Fiore-Donno, Bonkowski and Rose2018; Schlaeppi et al., Reference Schlaeppi, Dombrowski, Oter, Ver Loren van Themaat and Schulze-Lefert2014). In contrast to observations in cycads and hornworts, high host specificity and less influence by environmental factors were observed for cyanobacterial communities associated with feathermosses (Ininbergs et al., Reference Ininbergs, Bay, Rasmussen, Wardl and Nisson2011). Yet, in the lab, symbiotically competent Nostoc isolates from other hosts were generally capable to form a symbiotic association with these mosses (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). Another symbiosis that differs in terms of specificity is that of Azolla with T. azollae. This symbiosis is obligate and exists for more than 60 my (Carrapiço, Reference Carrapiço2006; Reference Carrapiço2010; Collinson, Reference Collinson2002). The obligatory nature is likely due to the symbiont’s highly reduced and eroded genome (Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010). Concomitantly, genomic sequencing and fingerprinting studies highlighted that this cyanobiont is co-evolving with its host (Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018; Zheng et al., Reference Zheng, Nilsson, Bergman and Rasmussen1999).
2.2 Genomics of symbiotically competent cyanobacteria
Genomes of several symbiotic cyanobacteria have been sequenced. Most illuminating among these were the genomes of obligate symbionts. Data on T. azolla and the nitrogen-fixing cyanobionts of the diatoms Rhopalodia gibba, UCYN-A (a symbiont of a haptophyte) and Epithemia turgida all point towards genome erosion (Kneip et al., Reference Kneip, Voβ, Lockhart and Maier2008; Nakayama et al., Reference Nakayama, Kamikawa, Tanifuji, Kashiyama, Ohkouchi, Archibald and Inagaki2014; Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010). All of these genomes are in the process of being streamlined while maintaining genes required for nitrogen fixation (N2-fixation). Epithemia turgida stands out by having completely eroded—or lost—genes for both photosystems.
In addition to the genome of T. azollae from Azolla filiculoides (Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010), cyanobiont genomes from five other Azolla species have been generated (Dijkhuizen et al., Reference Dijkhuizen, Brouwer, Bolhuis, Reichart, Koppers, Huettel, Bolger, Li, Cheng, Liu, Wong, Pryer, Weber, Bräutigam and Schluepmann2018; Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018). The first genome of a cyanobacterial symbiont dwelling in plants was that of Nostoc punctiforme isolated from a cycad (Meeks et al., Reference Meeks, Elhai, Thiel, Potts, Larimer, Lamerdin, Predki and Atlas2001). In contrast to that of T. azollae (5.49 Mb), its genome is 9.06 Mb and considered large for cyanobacteria (Meeks et al. Reference Meeks, Elhai, Thiel, Potts, Larimer, Lamerdin, Predki and Atlas2001; Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010). After that genome sequences from nine isolates from feathermosses, Blasia pusilla and one nonsymbiotic Nostoc strain CALU 996 have been reported (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017; Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018), followed by 4 fully assembled genomes of isolates from Blasia and hornworts (Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019), which are in the range of approximately 7 Mb in size, 6 isolates from a cycad (Gutiérrez-García et al., Reference Gutiérrez-García, Bustos-Díaz, Corona-Gómez, Ramos-Aboites, Sélem-Mojica, Cruz-Morales, Pérez-Farrera, Barona-Gómez and Cibrián-Jaramillo2019) and 10 isolates of facultative symbionts from Azolla (Pratte & Thiel, Reference Pratte and Thiel2021). For a comparison of genome sizes of cyanobacterial symbionts, see Figure 4a. In the next section, we will compare the findings gleaned from the different cyanobacterial genomes.
2.3 Comparative genomics: cyanobionts versus nonsymbiotic cyanobacteria
What is special about certain cyanobacteria from the order Nostocales that they are recurrently associated in symbioses? Phylogenetic analysis showed that all analysed cyanobacteria that occur in plant symbioses are included in a larger monophyletic clade that, however, also contains nonsymbiotic cyanobacteria (Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). While Nostoc isolates that can associate intracellularly with Gunnera do form a monophyletic clade, extracellular and epiphytic isolates are polyphyletic within the larger clade of symbionts (Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). A study by Nelson et al. (Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019), which sequenced genomes of additional isolates from bryophytes, corroborated this result. This further agrees with a phylogenomic survey across 650 diverse marine, freshwater and terrestrial cyanobacteria, that concluded that cyanobacteria with similar lifestyles tend to be more closely related to each other (Chen et al., Reference Chen, Teng, Zhao, Hu, Zhou, Han, Song and Shu2021).
2.3.1 Genomics of N2-fixation
The major contribution of the cyanobiont to the symbiosis with their plant hosts is fixed nitrogen. Filamentous cyanobacteria form specialised cells, called heterocysts, that have thicker cell walls, are photosynthetically inactive and create a microoxic environment inside the cell (Kumar et al., Reference Kumar, Mella-Herrera and Golden2010; Figure 3). In this oxygen-poor environment, high levels of oxygen-sensible nitrogenase occur (see Gallon, Reference Gallon1981). In plant–cyanobacterial symbioses, heterocyst formation and the rate of N2-fixation are increased; by how much depends on the respective host (Grilli Caiola et al., Reference Grilli Caiola, Canini and Moscone1989; Hill, Reference Hill1975; Wang et al., Reference Wang, Ekman and Bergman2004).

Figure 3. Syntrophic currencies exchanged by plant hosts and cyanobionts. A schematic Nostocales filament, consisting of vegetative cells and a heterocyst, is shown. The plant provides sugars (pink font), and the cyanobionts fix atmospheric nitrogen and provide it to the host via export (orange font). For an interesting discussion on putative transporters, see Roy et al. (Reference Roy, Reinders, Ward and McDonald2020).
N2-fixing cyanobacteria encode two types of nitrogenase. One nitrogenase requires molybdenum (Mo) and iron (Fe) as co-factors and is expressed only in heterocysts (Mo-nitrogenase; Thiel et al., Reference Thiel, Lyons and Erker1997). In the symbioses between T. azollae and A. filiculoides, it was hypothesised that Mo is provided and regulated by the host (Roy et al., Reference Roy, Reinders, Ward and McDonald2020). This is based on the observation that the genome of A. filiculoides encodes a Mo transporter (Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018), which, however, may likely be an importer for the plant (Roy et al., Reference Roy, Reinders, Ward and McDonald2020). Its upregulation under nitrogen deprivation hints to an involvement in symbiosis (Eily et al., Reference Eily, Pryer and Li2019). The Mo-nitrogenase is a complex that requires a range of enzymes for its synthesis and assembly (Dean et al., Reference Dean, Bolin and Zheng1993). The complex itself requires the tetraheteromeric dinitrogenase encoded by nifD and nifK and the homodimeric dinitrogenase reductase encoded by nifH. Several nitrogen-fixing cyanobacteria encode an additional vanadium-based nitrogenase, which also requires Fe (V-nitrogenase; Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019; Pratte & Thiel, Reference Pratte and Thiel2021; Thiel & Pratte, Reference Thiel and Pratte2001). In Anabaena variabilis (now N. variabilis), transcription of the V-nitrogenase is repressed by both fixed nitrogen and the presence of Mo (Pratte et al., Reference Pratte, Sheridan, James and Thiel2013). In the absence of Mo, V-nitrogenase activity is induced (Thiel, Reference Thiel1996). The gene cluster encoding V-nitrogenase and its regulators (Pratte et al., Reference Pratte, Sheridan, James and Thiel2013), and the vanadate transporters (Pratte & Thiel, Reference Pratte and Thiel2006), is conserved across cyanobacteria (Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019). The first two assembled genomes from plant cyanobionts (T. azollae and N. punctiforme PCC 73102), however, likely lack V-nitrogenase-encoding genes (Meeks et al., Reference Meeks, Elhai, Thiel, Potts, Larimer, Lamerdin, Predki and Atlas2001; Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010). Yet, recent studies (Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019; Pratte & Thiel, Reference Pratte and Thiel2021) report their presence in genomes of facultative, symbiotically competent cyanobacteria from Azolla, Blasia and hornworts. Nelson et al. (Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019) reported the gene cluster in two of the cyanobionts they studied located on a plasmid and suggested the option for a gain via horizontal gene transfer. If and when V-nitrogenase plays a role under Mo-limited conditions during cyanobacteria–plant symbioses needs to be further investigated. A recent study on V-nitrogenase activity in lichen-associated cyanobacteria showed that availability of Mo correlated with the rate of N2-fixation from V-nitrogenase (Darnajoux et al., Reference Darnajoux, Magain, Renaudin, Lutzoni, Bellenger and Zhang2019).
Most proteomic and transcriptomic approaches have focused on Mo-nitrogenase. Ekman et al. (Reference Ekman, Tollbäck and Bergman2008) provided a proteomic comparison between the cyanobiont of the floating fern A. filiculoides and the symbiotic Nostoc isolate N. punctiforme PCC 73102 grown in culture. These data show that in the symbiotic association of Azolla proteins related to nitrogen metabolism were enhanced while those related to photosynthesis were reduced in the cyanobiont compared to the free-living, symbiotically capable N. punctiforme isolate. Similar tendencies were observed in the proteomic comparison of N. punctiforme PCC 73102 in symbiosis with Gunnera manicata and in culture (Ekman et al., Reference Ekman, Tollbäck, Klint and Bergman2006). In contrast, the epiphytic associations appear different. In feathermosses, nitrogen is transferred from cyanobiont to host (Bay et al., Reference Bay, Nahar, Oubre, Whitehouse, Wardle, Zackrisson, Nilsson and Rasmussen2013), yet the expression of genes involved in N2-fixation or heterocyst formation is not induced in symbiotic versus free-living Nostoc (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). This suggests that the Nostoc isolates may have a similar rate of N2-fixation in their symbiotic and free-living state. In agreement with that, both symbiotically competent Nostoc isolates analysed in this study identified nifBHKU—that is, genes encoding structural components nifH and K, a factor involved in nitrogenase assembly (nifU; Dos Santos et al., Reference Dos Santos, Smith, Frazzon, Cash, Johnson and Dean2004; Johnson et al., Reference Johnson, Dos Santos and Dean2005) and an enzyme required for the FeMo co-factor assembly (nifB; Curatti et al., Reference Curatti, Ludden and Rubio2006)—and the regulator for heterocyst formation, hetR, in the top 10% of transcripts with the highest abundance in free-living and symbiotic condition (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). In summary, while in close association the symbiosis appears to lead to an induction of N2-fixation, the example of lose associations from the feathermosses suggests that here the host thrives already on the steady-state levels of fixed nitrogen provided by the cyanobionts. One could wonder whether this may be due to the specificity in symbiont recruitment described based on genomic fingerprinting of nifH sequences (Ininbergs et al., Reference Ininbergs, Bay, Rasmussen, Wardl and Nisson2011); yet the observations were likewise true for a moss-specific and the symbiotically competent N. punctiforme isolate (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017).
What makes cyanobacteria symbiotically competent? And do they all use similar mechanisms for associating with their hosts? The latter appears likely given that most of them—with the exception of the obligate symbiont T. azollae–are able to colonise different hosts (e.g., Pratte & Thiel, Reference Pratte and Thiel2021; Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). As such the question may rather be ‘What are the molecular mechanisms of cyanobacterial symbioses?’. To answer these two questions, (comparative) genomic, transcriptomic and proteomic analyses have been carried out in different host-cyanobacteria associations.
2.3.2 Cyanobacterial symbioses from colonisation to communication during a functional symbiosis
The ability to infect plants and associate in symbiosis is found in several members of the genus Nostoc/Anabaena. The last common ancestor of the monophyletic intra/extracellular symbiotic clade may have already been primed for the ability to associate with diverse hosts (Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). Yet this particular clade includes not only symbiotically competent isolates but also a variety of isolates not found in symbiosis. Warshan et al. (Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018) provide a set of 170 gene families that are exclusively encoded in the genomes of 10 facultative Nostoc symbionts. These include, among others, genes coding for transporters involved in the import of organic sulphur compounds, phosphate and branched amino acids and ammonium (Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). Genomic data on cycad-derived symbionts found several metabolite biosynthesis pathways specific to the symbiotic cyanobacteria (Gutiérrez-García et al., Reference Gutiérrez-García, Bustos-Díaz, Corona-Gómez, Ramos-Aboites, Sélem-Mojica, Cruz-Morales, Pérez-Farrera, Barona-Gómez and Cibrián-Jaramillo2019). Pratte and Thiel (Reference Pratte and Thiel2021) confirmed the presence of the majority of the gene families identified by Warshan et al. (Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018) in the genomes of 10 Nostoc isolates that had previously been isolated from Azolla (Meeks et al., Reference Meeks1988; Zimmerman et al., Reference Zimmerman, Rosen and Lumpkin1989). These isolates had originally been obtained as the putative permanent and vertically transferred cyanobiont of Azolla, but were later identified as cyanobacteria that colonised the cavity in addition to T. azollae (Papaefthimiou et al., Reference Papaefthimiou, Van Hove, Lejeune, Rasmussen and Wilmotte2008). Infection assays with Blasia sp. confirmed that these isolates are capable of associating in symbioses and suggest a wider host range than only Azolla (Pratte & Thiel, Reference Pratte and Thiel2021). However, genomic analyses of these symbiotic isolates and strains previously described as free-living identified several ‘symbioses genes’ in the free-living cyanobacteria. This may have two reasons: (a) Warshan et al. (Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018) used one particular cyanobacterial strain as nonsymbiotic control, while Pratte and Thiel used a diverse set; and (b) symbiotic compatibility may be present even in free-living isolates that currently have not been found in metagenomic studies of symbiotic communities.
As described above, the genome of the symbiont of Azolla is smaller than that of most other symbiotically competent cyanobacteria (Meeks et al., Reference Meeks, Elhai, Thiel, Potts, Larimer, Lamerdin, Predki and Atlas2001; Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019; Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010; Figure 4a). For one, T. azollae does not belong to the genus Nostoc and has a distinct phylogenetic placement (Nelson et al., Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019; Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). Moreover, the genome shows strong evidence of erosion (Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010). Many genes have been lost from the genome of T. azollae or subjected to pseudogenisation (Ran et al., Reference Ran, Larsson, Vigil-Stenman, Nylander, Ininbergs, Zheng, Lapidus, Lowry, Robert Haselkorn and Bergman2010; Figure 4b,c). Accordingly, 28 gene families present in facultative symbionts have been lost in T. azollae (Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). Among those were genes involved in chemotaxis and in transport and metabolism of alkane and aliphatic sulfonate, underpinning the differences between obligate and facultative symbioses.

Figure 4. Genome characteristics of genomes from symbiotic and nonsymbiotic cyanobacteria and cyanobacterial host plants. (a) Genome size of several cyanobacteria shown to be symbiotic or free-living (blue, cyano) and all sequenced plant hosts for cyanobacteria (yellow, plants). (b) Number of annotated protein-coding genes in the genomes of cyanobacteria (blue, cyano) and cyanobacterial host plants (yellow, plants). (c) Number of pseudogenes described for the here included cyanobacterial genome data (for details, see description below). (d) Relative repeat content in the assembled plant genomes from plants symbiotically associating with cyanobacteria. To have a mixed set of cyanobacteria associating with plants and free-living, we included cyanobacteria previously sequenced and/or included in the comparative analyses by Pratte & Thiel (Reference Pratte and Thiel2021) and Nelson et al. (Reference Nelson, Hauser, Gudiño, Guadalupe, Meeks, Salazar Allen, Villarreal and Li2019). NCBI accessions have been derived from these publications, and actual data have been obtained from the National Center for Biotechnology Information (NCBI). For cyanobacterial hosts, all host species that have been sequenced (Anthoceros agrestis, Anthoceros punctatus, Anthoceros angustus, Azolla filiculoides, Sphagnum fallax and Pleurozium schreberi) are included in the figure. The data are derived from Li et al. (Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018, Reference Li, Nishiyama, Waller, Frangedakis, Keller, Li, Fernandez-Pozo, Barker, Bennett, Blázquez, Cheng, Cuming, de Vries, de Vries, Delaux, Diop, Harrison, Hauser, Hernández-García and Szövényi2020), Pederson et al. (Reference Pederson, Warshan and Rasmussen2019), Zhang et al. (Reference Zhang, Fu, Li, Zhao, Liu, Li, Zwaenepoel, Ma, Goffinet, Guan, Xue, Liao, Wang, Wang, Wang, Zhang, Wang, Jia, Wang and Chen2020), and Genome statistic information from Phytozome genome ID: 522. Data on repeat content were not reported for S. fallax, and the number of pseudogenes has not been reported for cyanobacteria isolated from feathermosses.
2.3.2.1 Insights into motility and chemotaxis
What all cyanobionts have in common is their motility and ability to move towards their hosts. This is likewise true for facultative and obligate cyanobionts. In the case of Azolla, the filamentous motile stage—hormogonia—is induced to infect newly developed leaf cavities and sporocarps of the fern (Zheng et al., Reference Zheng, Rang and Bergman2009b). Hosts can induce the formation of hormogonia—as is discussed later in the section on the hosts. Yet, the ability to move is not all; extracts of diverse hosts are able to attract cyanobacteria chemotactically (Nilsson et al., Reference Nilsson, Rasmussen and Bergman2006). Genes involved in chemotaxis of hormogonia (cheA, B, D, R and W) are present in the genome of the symbiotically competent N. punctiforme (Meeks et al., Reference Meeks, Elhai, Thiel, Potts, Larimer, Lamerdin, Predki and Atlas2001). The absence of functional CheR methyltransferase results in loss of motility and reduced colonisation of N. punctiforme of Blasia (Duggan et al., Reference Duggan, Thiel and Adams2013). Furthermore, symbiotically competent strains induce chemotaxis-associated genes upon chemical contact with a putative host (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). Accordingly, genes relevant for chemotaxis are among the 170 symbioses genes recovered by Warshan et al. (Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). In contrast, among the genes lost from the genome of T. azollae are some families involved in chemotaxis (Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018). That is interesting because one would expect some host-derived guidance to the sporocarps or a new leaf cavity. But in contrast to facultative symbioses, movement of T. azollae may not only be guided by chemical communication. Trichomes are suggested to act as physical guidance towards new leaf cavities and sporocarps (Calvert et al., Reference Calvert, Pence and Peters1985; Calvert & Peters, Reference Calvert and Peters1981; Hill, Reference Hill1989). Nonetheless, metabolic-based communication may be altered during the initiation of sporocarp formation. The transcriptional profile of transporter-encoding genes from T. azollae changed significantly during the induction of sporocarps (Dijkhuizen et al., Reference Dijkhuizen, Tabatabaei, Brouwer, Rijken, Buijs, Güngör and Schluepmann2021). Already early on in research on Azolla, it was suspected that host-derived phenolics and flavonoids in trichomes may be involved in the movement of T. azollae (Carrapiço & Tavares, Reference Carrapiço and Tavares1989; Ishikura, Reference Ishikura1982; Pereira & Carrapiço, Reference Pereira and Carrapiço2007). Once the hosts—or new host tissue in case of Azolla—are colonised, hormogonia revert back to vegetative filaments (Bay et al., Reference Bay, Nahar, Oubre, Whitehouse, Wardle, Zackrisson, Nilsson and Rasmussen2013; Meeks & Elhai, Reference Meeks and Elhai2002; Zheng, Bergman, et al., Reference Zheng, Bergman, Chen, Zheng, Xiang and Rasmussen2009a). This repression is guided by the host and its regulation is governed by proteins encoded by the hrm-locus (Adams & Duggan, Reference Adams and Duggan2012 ; Campbell et al., Reference Campbell, Wong and Meeks2003; Cohen & Meeks, Reference Cohen and Meeks1997; Meeks, Reference Meeks1998).
The broad host range of many cyanobacterial symbionts that is occasionally observed in nature and used in the lab suggests that either (a) the symbioses rely on a common mechanism despite the diverse partners, (b) symbiotically competent Nostoc evolved the ability to recognise and respond a vast variety of different signals or (c) both.
2.3.2.2 Insights into nutrient exchange and transport
An important aspect of cyanobacterial biology during symbioses is its transporter system. In symbioses, photosynthesis is reduced in cyanobionts as a trade-off for the higher rates of N2-fixation (Bergman et al., Reference Bergman, Rai, Johansson and Söderbäck1992; Ekman et al., Reference Ekman, Tollbäck and Bergman2008; Peters & Meeks, Reference Peters and Meeks1989; Rai et al., Reference Rai, Borthakur, Singh and Bergman1989; Reference Rai, Soderback and Bergman2000). Thus, cyanobionts need to be provided with carbon from their hosts (Peters & Meeks, Reference Peters and Meeks1989; Rai et al., Reference Rai, Soderback and Bergman2000; Söderbäck & Bergman, Reference Söderbäck and Bergman1993; Stewart & Rodgers, Reference Stewart and Rodgers1977; Figure 3). Indeed, proteomics of T. azollae indicated an increase in a transporter associated—by sequence homology—with a phosphotransferase system fructose-specific IIC component (Ekman et al., Reference Ekman, Tollbäck and Bergman2008). In agreement, both in symbiosis with Gunnera and Azolla, key enzymes of the oxidative pentose phosphate pathway are induced (Ekman et al., Reference Ekman, Tollbäck, Klint and Bergman2006; Reference Ekman, Tollbäck and Bergman2008). This may suggest that fructose is the sugar transported between hosts and cyanobionts. However, a mutant study on sugar transporters of symbiotically competent N. punctiforme showed that only mutants where glucose permease function was impaired were unable to infect the hornwort Anthoceros punctatus (Ekman et al., Reference Ekman, Picossi, Champbell, Meeks and Flores2013). This leaves four options: (a) cyanobionts do not receive the same type of sugar from every host system, (b) protein sequences that were by homology associated with fructose transport and metabolism may need to be reinvestigated with new additional data at hand, (c) a lack of fructose during an established interaction may be circumvented, while glucose is required as a signal for the establishment of infection or (d) glucose is the transported sugar used as a carbon source for cyanobionts.
Warshan et al. (Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018) found that the genomes of facultative symbiotic cyanobacteria are enriched in transporters for phosphate and organic sulphur and the metabolism of the latter. Both phosphate and sulphate transporters are induced upon contact between the feathermoss Pleurozium schreberi and symbiotic Nostoc isolates (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). These data point to organic sulphur and phosphate as important nutrients delivered by the host. And indeed, sulphur compounds are a currency in feathermoss–cyanobiont symbioses (Stuart et al., Reference Stuart, Pederson, Weyman, Weber, Rassmussen and Dupont2020). Furthermore, elimination of a functional alkane sulfonate monooxygenase by targeted mutagenesis of symbiotic N. punctiforme showed that sulphur metabolism is required for establishment of a symbiosis with feathermosses (Stuart et al., Reference Stuart, Pederson, Weyman, Weber, Rassmussen and Dupont2020).
2.3.2.3 Insights into cyanobacterial stress during symbiosis
Nitrogen starvation induces a stress response in cyanobacteria (Christman et al., Reference Christman, Campbell and Meeks2011). It is thus not surprising that early proteomic analyses on the cyanobionts of Azolla and Gunnera identified an induction of proteins involved in stress (Ekman et al., Reference Ekman, Tollbäck, Klint and Bergman2006; Reference Ekman, Tollbäck and Bergman2008). In both host systems, these included proteins in oxidative stress. This agrees with data on cyanobacteria epiphytically associating with feathermosses. While both competent as well as non-competent Nostoc isolates highly expressed proteins involved in oxidative stress responses, some of these were only induced in the competent isolates upon physical contact with the mosses—both in the secreted proteome and transcriptome (Warshan et al., Reference Warshan, Espinoza, Stuart, Richter, Kim, Shapiro, Woyke, Kyrpides, Barry, Singan, Lindquist, Ansong, Purvine, Brewer, Weyman, Dupont and Rasmussen2017). Moreover, these same proteins were reduced in the secreted protein fraction of the non-competent Nostoc isolate in this study. Additionally, in a metabolic screen, some Nostoc isolates, including some cyanobionts from Blasia, have been described to produce the cyanobacterial toxin microcystin (Liaimer et al., Reference Liaimer, Jensen and Dittmann2016). A microcystin deficient mutant showed higher sensitivity to reactive oxygen species (ROS; Zilliges et al., Reference Zilliges, Kehr, Meissner, Ishida, Mikkat, Hagemann, Kaplan, Börner and Dittmann2011), suggesting a protective role of these compounds against oxidative stress. Indeed, the microcystins were not secreted by symbiotic Nostoc isolates supporting a role inside the symbiont (Liaimer et al., Reference Liaimer, Jensen and Dittmann2016). Overall, these data suggest that a vital part of establishing and possibly maintaining N2-fixing symbioses in many host-cyanobiont interactions relies on the induction of protection against ROS. This notion is corroborated by insights into a different nitrogen-fixing symbiosis: that occurring between rhizobial bacteria and legumes. A study by Günther et al. (2007) on Lotus japonicus has shown that high respiration rates mediated by leghemoglobins (which keep free O2 levels low while acting in internal O2 trafficking; see Appleby, Reference Appleby1984) lead to elevated levels of ROS.
2.4 Metagenomics and fingerprinting point to a bacterial microcosm surrounding the cyanobionts
No organism exists alone in its natural habitat. Already decades ago, the co-occurrence of other bacteria, next to cyanobionts, was noticed (Carrapiço, Reference Carrapiço1991; Wallace & Gates, Reference Wallace and Gates1986). Metagenome sequencing has uncovered a diversity of non-cyanobacterial symbionts (e.g., Dijkhuizen et al., Reference Dijkhuizen, Brouwer, Bolhuis, Reichart, Koppers, Huettel, Bolger, Li, Cheng, Liu, Wong, Pryer, Weber, Bräutigam and Schluepmann2018; Nelson et al., Reference Nelson, Hauser and Li2021). That cyanobionts are surrounded by various bacteria was noted for many systems. Thus, the host and cyanobiont do not only interact with each other, but with an entire microbial community. One might find this unsurprising. Bacteria and archaea are omnipresent, and a diversity of them associates with roots and leaves of plants. What roles they play and how essential the bacterial composition is to their plant hosts are, in general, the topics of current research (Fitzpatrick et al., Reference Fitzpatrick, Salas-González, Conway, Finkel, Gilbert, Russ, Teixeira and Dangl2020; Trivedi et al., Reference Trivedi, Leach, Tringe, Sa and Singh2020). The question though is, how are they capable to occupy the symbiotic space from the rather ‘open’ versions such as cavities of hornworts, liverworts and Azolla to more ‘closed’ options such as coralloid roots of cycads.
Are these bacteria capable of hijacking the symbiotic crosstalk between host and cyanobiont or do symbiotic spaces fulfil a general role in attracting symbionts for the plant? In a metagenomic study on hornwort cavities, it was observed that the nonsymbiotic associates follow the same patterns of specificity as the cyanobionts (Nelson et al., Reference Nelson, Hauser and Li2021). The original soil community dictates what organisms are present and as such have a chance to be recruited; here too, the diversity of the cavity does not equal that of the soil per se. Hence, Anthoceros has some measure of control, which microbes it allows in and which it does not. In Azolla species, there are recurring members of the microbial community that are attuned to the developmental status of the symbiotic organs and the cyanobacterial physiology (Dijkhuizen et al., Reference Dijkhuizen, Brouwer, Bolhuis, Reichart, Koppers, Huettel, Bolger, Li, Cheng, Liu, Wong, Pryer, Weber, Bräutigam and Schluepmann2018). These bacteria were found to be co-transferred during vegetative and sexual transmission of the cyanobacterial population (Carrapiço, Reference Carrapiço1991). In contrast to the other symbioses, in Gunnera petaloidea and Gunnera chilensis, fungi and bacteria were observed within both the gland cells and the mucilage (Johansson & Bergman, Reference Johansson and Bergman1992; Towata, Reference Towata1985), but there are no records on the presence of such microbes in the exact same symbiotic spaces as the cyanobionts. Yet, these are only microscopic observations, and no molecular insights have been garnered.
These observations suggest a complex interaction between host, cyanobacteria and other microorganisms. The nature of these interactions is to a large degree not yet fully understood. With the omics approaches at hand, the nature of these interactions can now be further investigated. Dijkhuizen et al. (Reference Dijkhuizen, Brouwer, Bolhuis, Reichart, Koppers, Huettel, Bolger, Li, Cheng, Liu, Wong, Pryer, Weber, Bräutigam and Schluepmann2018) performed the first comprehensive survey of the microbiome that occurs in six different Azolla species. The authors found that there are recurring Rhizobiales in the Azolla leaf pockets. Interestingly, similar presence of Rhizobiales has been reported in the endospheres of the hornwort Leiosporoceros (Bouchard et al., Reference Bouchard, Peñaloza-Bojacá, Toupin, Guadalupe, Gudiño, Salazar Allen, Li and Villarreal Aguilar2020), and the cycads Dioon and Zamia (Bell-Doyon et al., Reference Bell-Doyon, Laroche, Saltonstall and Villarreal Aguilar2020; Suárez-Moo et al., Reference Suárez-Moo, Vovides, Griffith, Barona-Gómez and Cibrián-Jaramillo2019). While these recent microbiome studies suggest that Rhizobiales might have certain significances in plant–cyanobacteria symbiosis, their prevalence and functional roles remain to be tested. Additionally, Dijkhuizen et al. (Reference Dijkhuizen, Brouwer, Bolhuis, Reichart, Koppers, Huettel, Bolger, Li, Cheng, Liu, Wong, Pryer, Weber, Bräutigam and Schluepmann2018) identified putative denitrifiers in the leave pocket of Azolla, suggesting that cheaters might exist in the symbiotic spaces. Yet, with the exception of the analyses of Azolla, no functional predictions have been made for the microbial community in cyanobacterial symbioses.
Currently, no comparative metagenomic study across the cyanobacterial hosts exists. Additionally, most metagenome sequencing studies focus on marker genes instead of entire genomes; this hampers inferences on functional roles in the symbiotic community and the identification of non-cyanobacterial symbionts or cheaters. Thus, how similar the microbiome in the symbiotic space of the phylogenetically distinct hosts is compared to nonsymbiotic tissues can only be approximated. Yet, because in all cyanobacterial hosts, facultative cyanobionts have some similar, symbiosis-associated genomic content and they perform similar functions within the symbiosis (Pratte & Thiel, Reference Pratte and Thiel2021, Warshan et al., Reference Warshan, Liaimer, Pederson, Kim, Shapiro, Woyke, Altermark, Pawlowski, Weyman, Dupont and Rasmussen2018), they might create a similar milieu in different symbiotic spaces. One could, hence, expect that the functional composition of the microbiome, be it beneficial or cheating, might be similar. Indeed, with more metagenomic data accumulating, it is becoming clear that while taxonomic compositions vary strongly, there are recurrent patterns in the functional composition of a microbiome in similar environments (Burke et al., Reference Burke, Steinberg, Rusch, Kjelleberg and Thomas2011). After all, it is the biochemical properties of an environment on which selection acts (see excellent discussion in Doolittle & Inkpen, Reference Doolittle and Inkpen2018). Future studies will illuminate convergence of functions in the symbiotic microverse across these diverse interactions.
3. Recurrent molecular patterns in cyanobacterial symbioses—a host-oriented perspective
The sequencing of cyanobacteria associated with nitrogen-fixing symbioses is for a long time on-going, and as a consequence, many of the strains have been sequenced. For practical reasons regarding the much larger genomes (Figure 4a), from cost to computational power and the availability of methods to cope with a high repetitive content (Figure 4d), the sequencing of the host genomes has progressed more slowly. However, in the last few years, the genomes of several host plants have been sequenced: Azolla filiculoides (Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018), four hornwort genomes [two Anthoceros agrestis ecotypes and A. punctatus (Li et al., Reference Li, Nishiyama, Waller, Frangedakis, Keller, Li, Fernandez-Pozo, Barker, Bennett, Blázquez, Cheng, Cuming, de Vries, de Vries, Delaux, Diop, Harrison, Hauser, Hernández-García and Szövényi2020) and Anthoceros angustus (Zhang et al., Reference Zhang, Fu, Li, Zhao, Liu, Li, Zwaenepoel, Ma, Goffinet, Guan, Xue, Liao, Wang, Wang, Wang, Zhang, Wang, Jia, Wang and Chen2020)] and the feathermoss P. schreberi (Pederson et al., Reference Pederson, Warshan and Rasmussen2019). Additionally, the 1KP project (Matasci et al., Reference Matasci, Hung, Yan, Carpenter, Wickett, Mirarab, Nguyen, Warnow, Ayyampalayam, Barker, Burleigh, Gitzendanner, Wafula, Der, dePamphilis, Roure, Philippe, Ruhfel, Miles and Wong2014) covers transcriptomes of G. manicata, 4 cycads (Cycas micholoitzii, Stangeria eriopus, Dioon edule and Encephalartos barteri), 10 different hornwort species from diverse genera, the liverwort Blasia sp. and an additional Azolla species. Additional transcriptomes for A. filiculoides, two other Azolla species, Azolla pinnata and Azolla caroliniana, and the hornwort A. punctatus exist (Brouwer et al., Reference Brouwer, Bräutigam, Buijs, Tazelaar, van der Werf, Schlüter, Reichart, Bolger, Usadel, Weber and Schluepmann2017; Chatterjee et al., Reference Chatterjee, Schafran, Li and Meeks2021; de Vries et al., Reference de Vries, Fischer, Roettger, Rommel, Schluepmann, Bräutigam, Carlsbecker and Gould2016; Li et al., Reference Li, Nishiyama, Waller, Frangedakis, Keller, Li, Fernandez-Pozo, Barker, Bennett, Blázquez, Cheng, Cuming, de Vries, de Vries, Delaux, Diop, Harrison, Hauser, Hernández-García and Szövényi2020; Qi et al., Reference Qi, Kuo, Guo, Li, Li, Qi, Wang, Hu, Xiang, Zhang, Guo, Huang and Ma2018).
3.1. Establishment of interaction with cyanobacterial and other symbionts
Some of the now existing data were already available and included in a large comparative analysis that targeted the distribution and evolution of the symbiotic toolkit associated with arbuscular mycorrhizal (AM) symbioses (Radhakrishnan et al., Reference Radhakrishnan, Keller, Rich, Vernié, Mbadinga Mbadinga, Vigneron, Cottret, San Clemente, Libourel, Cheema, Linde, Eklund, Shifeng Cheng, Wong, Lagercrantz, Li, Oldroyd and Delaux2020). The authors found that symbiotic genes involved in these particular symbioses are not present in land plants undergoing cyanobacterial but no AM symbioses. This suggests that even though the symbiotic pathways associated with the establishment of AM symbioses have been recruited for nitrogen-fixing symbioses with rhizobia (Parniske, Reference Parniske2008), cyanobacterial symbioses rely on different pathways for their interaction. Several data from Gunnera, cycads, hornworts, Blasia and feathermosses show that specific attractants exist (Bay et al., Reference Bay, Nahar, Oubre, Whitehouse, Wardle, Zackrisson, Nilsson and Rasmussen2013; Campbell & Meeks, Reference Campbell and Meeks1989; Khamar et al., Reference Khamar, Breathwaite, Prasse, Fraley, Secor, Chibane, Elhai and Chiu2010; Nilsson et al., Reference Nilsson, Rasmussen and Bergman2006; Rasmussen et al., Reference Rasmussen, Johansson and Bergman1994), and that the presence of the hosts induces hormogonia formation (Bay et al., Reference Bay, Nahar, Oubre, Whitehouse, Wardle, Zackrisson, Nilsson and Rasmussen2013; Campbell & Meeks, Reference Campbell and Meeks1989; Meeks & Elhai, Reference Meeks and Elhai2002; Ow et al., Reference Ow, Gantar and Elhai1999; Rasmussen et al., Reference Rasmussen, Johansson and Bergman1994; Figure 1). Even in the interaction between Azolla and its cyanobiont, hormogonia formation and specific attraction to the sporocarps as well as the apex of the shoot (where new leaves are formed) is observed (Calvert et al., Reference Calvert, Pence and Peters1985; Hill, Reference Hill1989; Perkins & Peters, Reference Perkins and Peters1993; Peters et al., Reference Peters, Toia, Raveed and Levine1978; Peters & Meeks, Reference Peters and Meeks1989; Zheng, Bergman, et al., Reference Zheng, Bergman, Chen, Zheng, Xiang and Rasmussen2009a).
Campbell and Meeks (Reference Campbell and Meeks1989) found a hormogonia inducing factor (HIF) of a size between 12 and 14 kDa in the hornwort A. punctatus. They did not further identify the nature of the secreted signal but suggested a poly-phenol-like molecule (Campbell & Meeks, Reference Campbell and Meeks1989; Meeks & Elhai, Reference Meeks and Elhai2002). Rasmussen et al. (Reference Rasmussen, Johansson and Bergman1994) identified that mucilage of different Gunnera species induced hormogonia. Here, too, a molecule of about 12 kDa was found. In this case, proteinase K treatment suggested it to be a peptide. The accumulating transcriptomic and genomic data can now be helpful in identifying short peptides that may act as HIFs and test their function in the lab. Another HIF, a mixture of diacylglycerols, was recently identified from the cycad Cycas revoluta (Hashidoko et al., Reference Hashidoko, Nishizuka, Tanaka, Murata, Murai and Hashimoto2019). The major compound 1-palmitoyl-2-linoleoyl-sn-glycerol was the most active HIF. Additionally, sugars are in discussion as putative HIFs. Indeed, the secreted mucilage of mature glands of Gunnera and soluble sugars within it induced hormogonia formation (Khamar et al., Reference Khamar, Breathwaite, Prasse, Fraley, Secor, Chibane, Elhai and Chiu2010).
Because N2-fixation is inhibited in hormogonia, a reversion to filaments with heterocysts, the N2-fixing cells of cyanobacteria, is needed for the establishment of successful symbioses. This is triggered by all cyanobacterial symbioses (Meeks & Elhai, Reference Meeks and Elhai2002). Glucose and fructose, which are almost absent from the mucilage of Gunnera, but accumulate within the gland tissues, repressed the formation of hormogonia (Khamar et al., Reference Khamar, Breathwaite, Prasse, Fraley, Secor, Chibane, Elhai and Chiu2010). Sugars have also been highlighted as putative attractants for the cyanobionts (Nilsson et al., Reference Nilsson, Rasmussen and Bergman2006). Functional analyses of the genome of the symbiotic N. punctiforme showed that it encodes a functional fructose transporter, a glucose permease and an OprB family porin, which was associated with the uptake of both sugars (Ekman et al., Reference Ekman, Picossi, Champbell, Meeks and Flores2013). In other cyanobacteria, this porin family has recently been associated with transport of different metals (Qiu et al., Reference Qiu, Jiang, Lis, Li, Deng, Shang, Sun, Keren and Qiu2021; Schätzle et al., Reference Schätzle, Brouwer, Liebhart, Stevanovic and Schleiff2021). Additionally, some mutants of porin-like genes from Anabaena sp. PCC 7120 had altered phenotypes of the outer membrane, affected integration of other porins in the membrane or affected nitrogen demand and/or fixation (Schätzle et al., Reference Schätzle, Brouwer, Liebhart, Stevanovic and Schleiff2021). Hence, the OprB family appears to have a diversity of roles in bacterial biology; however, whether the role of OprB in sugar uptake is primary or a secondary effect cannot be distinguished because membrane integrity or integration of other proteins into the membrane was not assessed by Ekman et al. in 2013. In agreement with the data from Khamar et al. (Reference Khamar, Breathwaite, Prasse, Fraley, Secor, Chibane, Elhai and Chiu2010), all mutants for the fructose and glucose transporters were able to form hormogonia (Ekman et al., Reference Ekman, Picossi, Champbell, Meeks and Flores2013). Yet only mutants unable to produce the glucose permease resulted in the abortion of infection of the hornwort A. punctatus (Ekman et al., Reference Ekman, Picossi, Champbell, Meeks and Flores2013). Given that the sugar content, including glucose, is depleted in Gunnera glands once successfully colonised by cyanobacteria (Khamar et al., Reference Khamar, Breathwaite, Prasse, Fraley, Secor, Chibane, Elhai and Chiu2010) suggests that glucose may additionally be supplemented by the plant as food for the symbiont.
3.2. Symbiotic communication and integration into stress responses
Transcriptomic analyses of the interaction between A. filiculoides and T. azollae recently highlighted a chalcone synthase-encoding gene (CHS) that has highly induced transcript levels only in the presence of the cyanobiont (Eily et al., Reference Eily, Pryer and Li2019; Li et al., Reference Li, Brouwer, Carretero-Paulet, Cheng, de Vries, Delaux, Eily, Koppers, Kuo, Li, Simenc, Small, Wafula, Angarita, Barker, Bräutigam, dePamphilis, Gould, Hosmani and Pryer2018), suggesting a commitment to flavonoid biosynthesis in symbiosis. And indeed, deoxyanthocyanins alone can increase the induction of the hormogonia-repressing factor hrmA by 70% and even more so in combination with naringenin, but not with other phenylpropanoid(-derived) compounds that were tested by Cohen et al. (Reference Cohen, Sakihama, Takagi, Ichiba and Yamasaki2002). The inhibiting effect was also visible when extracts of leaf tissue from different Azolla species were used (Cohen et al., Reference Cohen, Sakihama, Takagi, Ichiba and Yamasaki2002). Agreeingly, several flavonoids were unable to promote hormogonia formation (Campbell & Meeks, Reference Campbell and Meeks1989; Nilsson et al., Reference Nilsson, Rasmussen and Bergman2006). Nonetheless, phenolics, including flavonoids, have been suggested to be involved in the transfer of cyanobionts to newly developed leaf cavities and sporocarps of Azolla (Pereira & Carrapiço, Reference Pereira and Carrapiço2007). Expression of homologs of genes encoding enzymes in the phenylpropanoid pathway (see also de Vries et al., Reference de Vries, Fürst-Jansen, Irisarri, Dhabalia Ashok, Ischebeck, Feussner, Abreu, Petersen, Feussner and de Vries2021b), providing the precursors for flavonoid biosynthesis, and flavonoid biosynthesis in A. filiculoides, is dependent on the circadian rhythm and the availability of nitrogen (Güngör et al., Reference Güngör, Brouwer, Dijkhuizen, Shaffar, Nierop, de Vos, Toraño, van der Meer and Schluepmann2021). Anthocyanins produced by Azolla are elevated under various stresses, including protection against light stress (Nham Tran et al., Reference Nham Tran, Miranda, Abeynayake and Mouradov2020). It is no surprise that genes coding for enzymes involved in anthocyanin biosynthesis are responsive to various triggers and stresses. One of the intriguing questions is what role do they play in the symbiosis. Given the permanent relationship of Azolla and its cyanobiont that needs to be maintained during stress, it is conceivable that stress-responsive metabolites, such as flavonoids, act as signals in the Azolla spp.–Trichormus symbioses. Proteomic analyses of salt stress on Azolla microphylla showed changes affecting the plant physiology and enzymes involved in N2-fixation of the cyanobiont (Thagela et al., Reference Thagela, Yadav, Mishra, Dahuja, Ahmad, Singh, Tiwari and Abraham2017; Yadav et al., Reference Yadav, Tripathi, Mishra, Ramteke, Singh and Abraham2019). The phytohormone salicylic acid (SA), which is involved in biotic stress in land plants (Pieterse et al., Reference Pieterse, Leon-Reyes, Van der Ent and Van Wees2009), alters cyanobiont abundance and nifE gene expression (de Vries et al., Reference de Vries, de Vries, Teschke, von Dahlen, Rose and Gould2018). Vice versa, transcriptomics and comparative genomic analyses highlight that the presence or absence of the cyanobiont alters the expression of homologous genes putatively involved in SA biosynthesis and signalling (de Vries et al., Reference de Vries, Herrfurth, Li, Feussner and de Vries2021a). More experimental and comparative data are needed to understand (a) how stress-responsive signalling integrates into Azolla’s symbiosis and (b) whether this can be transferred to non-permanent cyanobacterial symbioses.
3.3. Nutrient exchange from a genomic host perspective
A recurrent theme when analysing large-scale data from cyanobacteria and their hosts is, as already exemplified above, the transport of sugars to the cyanobionts as a counter-currency of the symbioses (Figure 3). Indeed, carbon transport from feathermosses to their epiphytic cyanobionts was demonstrated (Stuart et al., Reference Stuart, Pederson, Weyman, Weber, Rassmussen and Dupont2020). Transcriptomic analyses of the hornworts A. punctatus and A. agrestis with and without cyanobiont highlighted a sugar transporter from the SWEET1 clade that increases its transcript level in a symbiotic state (Li et al., Reference Li, Nishiyama, Waller, Frangedakis, Keller, Li, Fernandez-Pozo, Barker, Bennett, Blázquez, Cheng, Cuming, de Vries, de Vries, Delaux, Diop, Harrison, Hauser, Hernández-García and Szövényi2020). In the genome of A. filiculoides, 15 different SWEET genes have been identified, none of which can be linked to the symbioses based on transcriptomic data from cyanobiont-containing and cyanobiont-free cultures (Eily et al., Reference Eily, Pryer and Li2019). That does not mean that they play no role in symbiosis in general; furthermore, other sugar transporters could be involved. This illustrates that the picture cannot be transferred between symbiotic systems in a 1:1 manner. Additionally, the case of A. filiculoides might be special because of the perpetuity of its symbiosis. Whether facultative associations between evolutionary distinct plants and their cyanobionts use similar mechanisms is to be tested. Yet, it is not unthinkable that convergent molecular mechanisms have emerged that tip into the same pathways, given that the cyanobacteria appear capable of infecting many of these possible hosts.
4. Conclusion
Cyanobacteria can occur as symbionts in a diversity of plants that are separated by millions of years of evolution. Despite this diversity, genomic data indicate that among the facultative symbionts, similar molecular mechanisms are used to interact with their hosts. This is in agreement with the ability of these cyanobionts to infect distinct host lineages. In contrast, transcriptomic data on the cyanobacteria and their hosts hint that every host system recruits unique molecular features particular to each individual association; this is in line with the convergent evolution of these cyanobacterial symbioses and the unique characters and different degrees of intimacy observed in each system. It is conceivable that symbiotically competent cyanobacteria must have some plasticity in their response to the different hosts and an ability to recognise each individual species. Comparative genomics and transcriptomics have the power to illuminate shared patterns across these diverse systems.
Acknowledgement
We thank the anonymous reviewer for several valuable comments.
Financial support
J.d.V. thanks the European Research Council for funding under the European Union’s Horizon 2020 research and innovation programme (Grant Agreement No. 852725; ERC-StG ‘TerreStriAL’). J.d.V. is grateful for support through the German Research Foundation (DFG) within the framework of the Priority Programme ‘MAdLand—Molecular Adaptation to Land: Plant Evolution to Change’ (SPP 2237; VR 132/4-1).
Conflicts of interest
The authors declare no conflicts of interest.
Authorship contributions
S.d.V. and J.d.V. have outlined and written the manuscript together; both S.d.V. and J.d.V. created the figures.
Data availability statement
Data availability is not applicable to this article as no new data were created or analysed in this study.








Comments
Dear Dr. Hamant,
Thank you for the invitation to submit our Review article "Evolutionary genomic insights into cyanobacterial symbioses in plants" to Quantitative Plant Biology.
As previously outlined, we discuss recent developments in the field of plant–cyanobacteria symbiosis, which were driven by the generation of high-throughput-data. We pinpoint their power to yield general patterns across these diverse symbioses occurring in representative plant species from across all major lineages.
Yours sincerely
Sophie de Vries and Jan de Vries