INTRODUCTION
Giardia intestinalis is a common cause of waterborne gastrointestinal illness and the most common human intestinal parasite identified by public health laboratories in the USA [Reference Kappus1]. Each year about 20 000 cases of giardiasis are reported in the USA, with most cases occurring sporadically; 1–2% of cases are associated with an outbreak [2]. The most common routes of transmission for sporadic giardiasis are person-to-person contact, participation in recreational water activities, and consuming untreated or treated but unfiltered surface water and shallow well water [Reference Birkhead and Vogt3–Reference Stuart8]. Outbreaks of giardiasis most frequently occur through person-to-person transmission in daycare centres and through waterborne transmission in households with a contaminated private water source. Since the Safe Drinking Water Act of 1974 and its subsequent 1986 and 1996 amendments, large outbreaks associated with community water sources are uncommon in the USA (Table 1) [9–13].
Table 1. Drinking-water-associated outbreaks, USA, 1997–2006
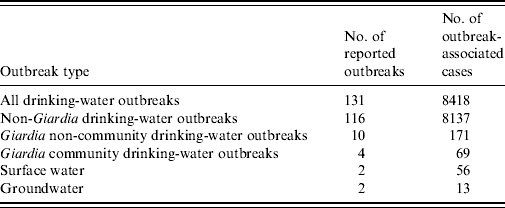
Community and non-community water systems are public water systems that have at least 15 service connections or serve an average of at least 25 residents for at least 60 days per year. A community water system serves year-round residents of a community, subdivision, or mobile home park. A non-community water system serves an institution, industry, camp, park, hotel, or business.
We report identification of an outbreak of giardiasis associated with a community drinking-water system and the resulting epidemiological, laboratory, and environmental investigation. This outbreak is noteworthy in that it was the largest outbreak of giardiasis associated with a community drinking-water source in the USA in 10 years. Furthermore, the outbreak was associated with groundwater, an infrequent source of parasitic disease outbreaks.
MATERIALS AND METHODS
Detection of the outbreak
On 9 September 2007, the New Hampshire Department of Environmental Services (NHDES) issued a boil water order for a 205-home community serviced by a common water system after routine water sampling detected the presence of total and faecal coliform bacteria within the distribution system. The community water system was divided into a west system (system A) and an east system (system B), with a number of wells supplying each (Fig. 1). Although the systems connected at one point through a valve, prior to and during the outbreak the valve was closed and the two systems were functionally separate, each with distinct water sources. On 17 September, NHDES received reports from two system A residents alleging infection with Giardia. NHDES notified the New Hampshire Department of Health and Human Services (NHDHHS) and an outbreak investigation was initiated.
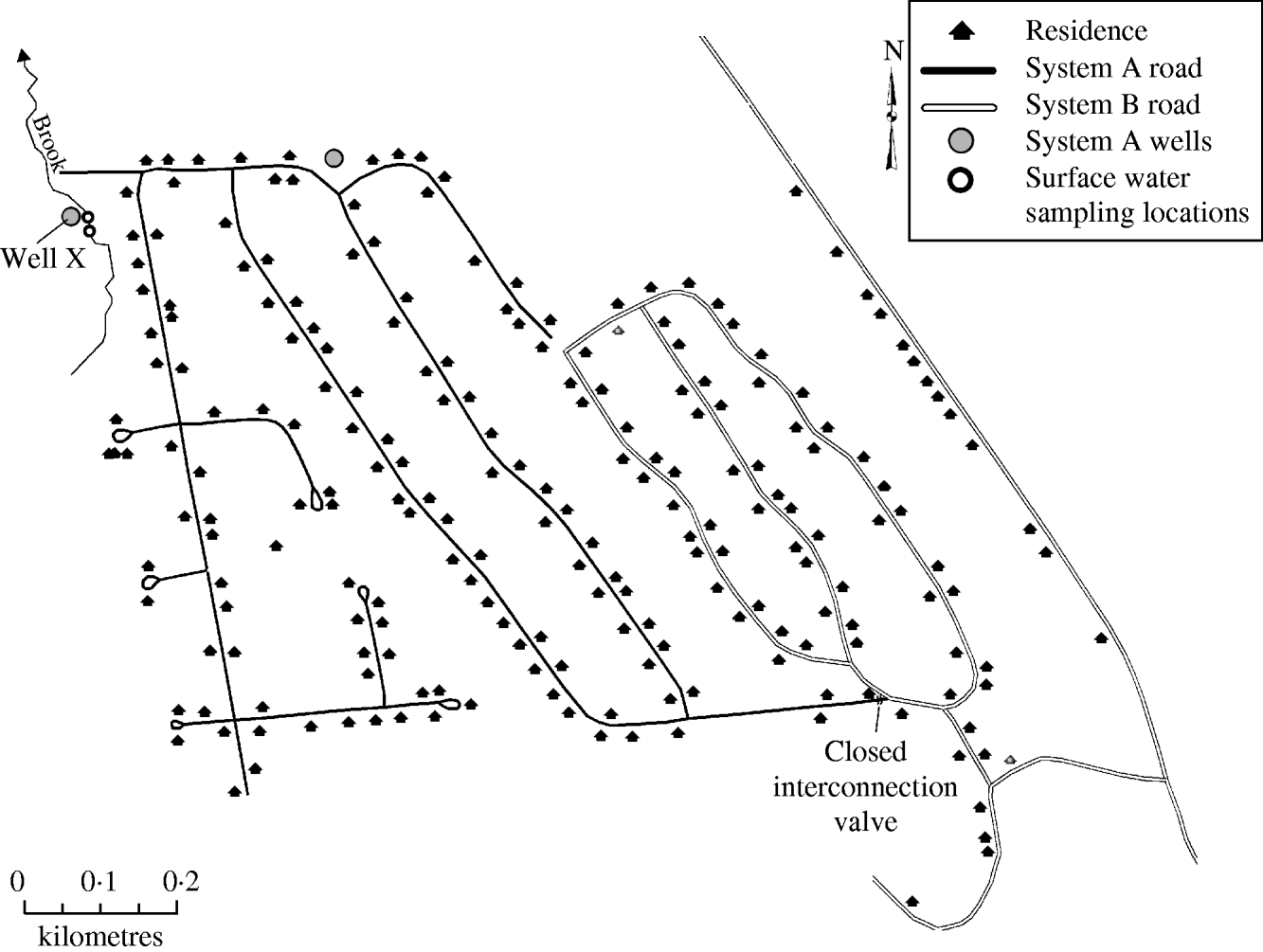
Fig. 1. Schematic of water system, giardiasis outbreak investigation, New Hampshire, 2007.
Case definition and case finding
Confirmed, probable, and suspect case definitions were developed. Confirmed cases were residents of system A or system B with a positive Giardia laboratory test result and onset of gastrointestinal illness on or after 15 August or if asymptomatic, specimen collection date on or after 15 August. Probable cases were residents who experienced diarrhoea (⩾3 loose stools in 24 h) on or after 15 August without a positive laboratory test and whose residence drew from the same water system as a confirmed case. Suspect cases were residents who experienced gastrointestinal symptoms other than diarrhoea on or after 15 August without a positive laboratory test and whose residence drew from the same water system as a confirmed case. Available communicable disease surveillance reports from the months of August and September were reviewed to identify potentially related giardiasis cases. Local hospital laboratories were asked to forward all Giardia specimens to NHDHHS for additional testing. A questionnaire administered as part of a cohort study asked household members about gastrointestinal illness in residents since 15 August.
Cohort study
A cohort study was conducted to identify risk factors for giardiasis. The cohort study questionnaire inquired about water consumption habits, use of water filtration devices, and details of gastrointestinal illness experienced since 15 August. Questionnaires were distributed to water system customers on 21 September and returned by mail between 22 September and 1 November. The survey was distributed to both system A and system B residents because water system distribution details were unclear at the time of survey distribution. Statistical analyses were conducted using SAS version 9.1 (SAS Institute Inc., USA). Univariate methods were used to evaluate attack rates of giardiasis for exposures and to calculate measures of association (risk ratios) between various water exposures and giardiasis. Corresponding 95% confidence intervals were used to assess statistical significance. A multivariate analysis using hierarchical backwards elimination of a logistic regression model was conducted to evaluate relationships between significant exposures identified in the bivariate analysis.
Environmental investigation
Following issuance of the boil order, NHDES staff and the water system operator conducted a sanitary survey of the water system to assess potential entry point(s) of bacterial contamination, which included inspections of water system pumping facilities, atmospheric storage tanks, piping access points, and water sources. Additionally, NHDHHS and NHDES staff conducted numerous field visits to the system to assess other factors that may have been related to the outbreak and to collect water and environmental samples.
Laboratory investigations
Human specimens
Stool specimens collected by private healthcare providers were sent to local hospital laboratories to undergo direct fluorescent antibody (FA) testing using the Merifluor Cryptosporidium/Giardia kit (Meridian Bioscience, USA) to identify Giardia. All available specimens were sent to NHDHHS for further testing. Additionally, NHDHHS offered residents free Giardia stool testing. Specimens sent to NHDHHS for FA testing were subsequently sent to the Centers for Disease Control and Prevention (CDC) for polymerase chain reaction (PCR) testing and subtyping. At CDC, after washing specimens twice in distilled water, genomic DNA was extracted from 0·2 ml of the washed faecal pellet using FastDNA Spin kit for soil (BIO 101, USA) according to the manufacturer's instructions. Giardia cysts present in the specimens were genotyped by nested PCR amplification of a 532-bp fragment of the triosephosphate isomerase (TPI) gene [Reference Sulaiman14]. All secondary PCR products were sequenced on an ABI3100 Genetic Analyzer (Applied Biosystems, USA) to identify the genotype and subtype present, using ABI BigDye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems). Each specimen was analysed at least twice by PCR. DNA of G. duodenalis assemblage D was used as the positive control in all TPI-based PCR analysis.
Home water filtration devices
The cohort study survey assessed water filtration devices used in the home. Respondents with home water filtration devices were contacted and asked to submit the filter for testing. One water pitcher filter (carbon filter) from a system A home was sent to CDC for Giardia testing using FA and differential interference contrast (DIC) microscopy and PCR. The housing of the filter was cut using a sterile scalpel and the carbon filter particles transferred to sterile bottles for elution of Giardia cysts. The laureth-12 eluent specified by United States Environmental Protection Agency (USEPA) Method 1623 [15] was used to elute the carbon filter particles using 1 min of hand shaking followed by 10 min on wrist action shaker. The samples were then allowed to settle, and the supernatant was poured through a 70-μm sterile nylon cell strainer into 50-ml tubes. A second elution of the carbon particles was performed using the same procedures. The filtrate from both elution steps was pooled in a 200-ml centrifuge tube and centrifuged at 4000 g for 30 min at 4°C. Supernatant was removed and the remaining pelleted material (∼1 ml of packed pellet) subjected to immunomagnetic separation (IMS) in two separate IMS reactions. Giardia cysts in one of the IMS suspensions were processed for FA and DIC microscopy by USEPA Method 1623 [but 4′,6-diamidino-2-phenylindole (DAPI) staining was not performed].
Giardia cysts in the other IMS suspension were used directly in DNA extraction for PCR without cyst detachment. DNA was extracted using the QIAamp DNA Mini kit (Qiagen, USA) after the cysts were subjected to five cycles of freeze-thaw (−70°C for 30 min and 56°C for 30 min) in 180 μl ATL buffer from the QIAamp kit, and overnight digestion in 1 mg/ml of proteinase K (Sigma, USA) at 56°C. Giardia genotypes and subtypes in the 100 μl of extracted DNA were determined by TPI–PCR and sequenced as described above. Each DNA preparation was analysed five times by PCR, using 2 μl of extracted DNA per PCR.
Water distribution samples
Water distribution samples from system A and system B were collected and tested for total and faecal coliforms on 6 September. Repeat samples were collected on 9 September and then regularly during the following 2 weeks until two consecutive samples collected at least 24 h apart were coliform free. Samples were analysed for total and faecal coliforms at a private laboratory.
Well water samples
Well water was collected from both wells that served system A on 9 September and tested for total and faecal coliforms by a private laboratory and NHDES. After faecal coliforms were identified in one well (well X) additional well X water samples, each ∼1000 l, were collected by the water operator on 18 and 24 September and sent to a private laboratory for Giardia testing using the USEPA Method 1623. A final well X water sample was collected on 7 November by NHDES for microscopic particulate analysis (MPA) for analysis and evaluation of the likelihood that the well was under the direct influence of surface water. The method included pumping ∼2140 l of groundwater from the well through a 1-μm, woven honeycomb filter over the course of an 8- to 12-h period at 3·8 l/min. Prior to passing discharge water through the filter, field screening of well discharge water was performed for pH, specific conductance, dissolved oxygen, and temperature over an approximate 14-h period to ensure that well-water chemistry stabilized prior to filter collection. The filter was then packaged and sent via standard chain of custody protocols by overnight courier to the USEPA Region 10 laboratory for MPA analysis by USEPA's 1992 Consensus Method for MPA Analyses [16]. A 72-l well X water sample was also sent to CDC for Giardia testing. The CDC processed the well-water sample using the ultrafiltration method of Hill et al. [Reference Hill17]. The pelleted material (∼0·5 ml) from the filter elution was split into two aliquots and each aliquot processed by IMS. Purified Giardia cysts from one of the IMS reactions were analysed by FA and DIC microscopy by USEPA Method 1623 [15] (but DAPI staining was not performed). Giardia cysts from the other IMS reaction were analysed by nucleic acid extraction and TPI–PCR [Reference Sulaiman14] as previously described for the materials eluted from the home water filtration device.
Surface water samples
A 34-l sample of surface water from the brook adjacent to well X was collected on 9 October and sent to CDC for Giardia testing. The sample was collected in two successive aliquots whereby the sampler waded into the surface water (brook) surrounding the well at two different locations and sequentially filled one half of the 40-l polyethylene cubitainer by successively dipping a 1-l HDPE grab sample bottle into the surface water and transferring water into the cubitainer with a plastic funnel. One sampling location was ∼25 m east of the well (side stream) and the other location ∼13 m south of the well (upstream). At CDC the sample was concentrated by ultrafiltration according to the method of Hill et al. [Reference Hill17] and centrifuged (resulting in a packed pellet of ∼1 ml), and tested for Giardia by IMS/FA and TPI–PCR as previously described for the materials eluted from the home water filtration device.
Beaver samples
Weather during the late autumn and winter prevented beaver trapping until the following spring, March 2008. Beavers from the surface water habitat were collected. Necropsies were performed, and faecal samples were obtained from the beavers within 24 h of death. Samples were sent to CDC for DNA extraction and Giardia testing by PCR, using the same techniques as in the analysis of human specimens.
RESULTS
Case definition and case finding
Thirty-one confirmed (n=17) and probable (n=14) cases affecting 27 (63%) of 43 responding system A households were identified; four suspect cases were also identified. No confirmed cases were identified in system B; therefore, no system B residents met the confirmed, probable or suspect case definitions. Of the 31 cases, 18 cases were identified through the cohort study, 12 were dually identified though routine communicable disease surveillance and the cohort study, and one was identified only through routine communicable disease surveillance. Four (15%) of 27 case households were home to two case-patients; in three of the four households, cases became ill several days apart and are potential secondary cases. The median age of cases was 49 years (range 3–83 years). Children aged <5 years accounted for one (3%) of 31 cases. Sixty-one per cent of cases were male. Patients had illness onsets from 20 August to 25 September (Fig. 2). Of 30 patients with clinical information, symptoms reported included diarrhoea (87%), abdominal cramps (83%), fever (23%), vomiting (23%), and bloody diarrhoea (10%). Reported duration of illness ranged from 2 to 30 days. Fourteen (54%) of 26 patients with diarrhoea reported experiencing recurrent diarrhoea, defined as diarrhoea that resolved for more than 1 day and later returned. Eighteen (60%) of 30 cases sought medical care. No deaths or illnesses requiring hospitalization were reported.
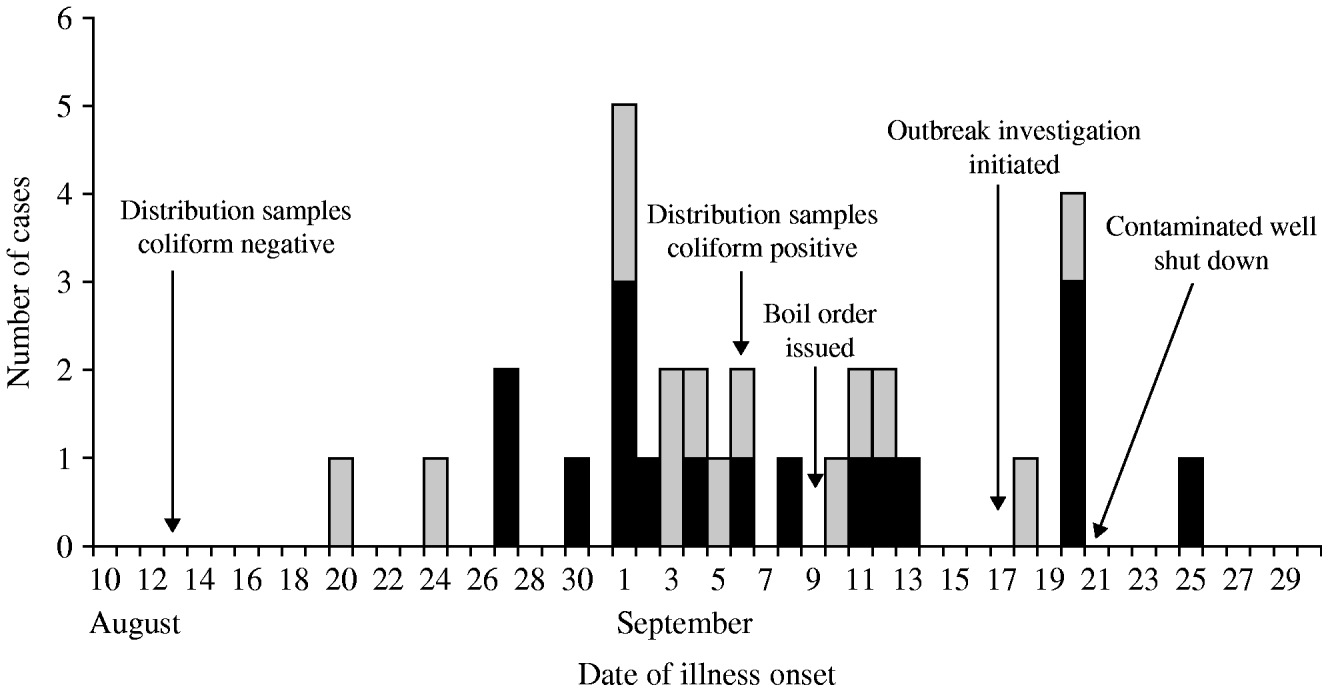
Fig. 2. Timeline of the outbreak, giardiasis outbreak investigation, New Hampshire, 2007. ▪, Confirmed cases (n=17); ![]() , probable cases (n=14).
, probable cases (n=14).
Cohort study
Two hundred surveys were distributed to system A and system B households. Sixty-two (31%) households returned completed surveys. Because no system B residents met the case definition, only residents of system A were included in the analysis. Forty-three (36%) of 128 system A households returned surveys, which included information on 100 individual residents. The four suspect cases were excluded from risk factor analysis and one laboratory-confirmed case refused participation in the cohort study. Consuming tap water was significantly associated with illness (Table 2). Of 63 individuals who consumed tap water, 27 (43%) reported illness compared with three (9%) of 33 who did not consume tap water [risk ratio (RR) 4·7, 95% confidence interval (CI) 1·5–14·4). Drinking four or more cups of tap water a day increased the risk of infection (RR 5·0, 95% CI 2·5–10·0), and a significant trend was detected between drinking increasing amounts of tap water and increased risk of infection (χ2 test for trend=28·9, P<0·001). Bottled water was a protective exposure (RR 0·4, 95% CI 0·2–0·9) in the univariate analysis since respondents who drank no tap water were 17·6 times more likely to drink bottled water (P value<0·001). Given the relationship between tap water and bottled water consumption, bottled water was no longer significantly protective in a multivariate logistic regression model in which only consuming tap water remained a significant risk factor for infection.
Table 2. Risk factors for illness in system A cohort study survey respondents (n=100), giardiasis outbreak investigation, New Hampshire, 2007
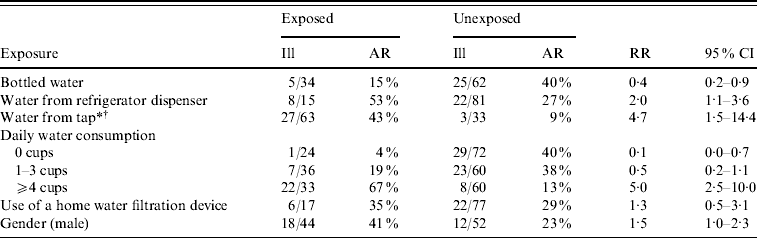
AR, Attack rate; RR, risk ratio; CI, confidence interval.
* χ2 test for trend=28·9, P<0·001.
† In a multivariate logistic regression model, only consuming tap water remained significantly associated with infection.
Environmental investigation
The 205-home community was rurally located in the northeastern part of New Hampshire. Sewer service was not provided to the community, and each home maintained their own on-lot septic system. The community drinking-water system was historically divided into a west (system A) and an east (system B) system (Fig. 1). Although the systems connected at one location through a closed valve, the two systems were functionally separate, each with distinct water sources prior to and during the outbreak. System A was served by two groundwater wells and system B was served by four groundwater wells. The water-supply well types were believed to be drilled in bedrock based on water screening parameters although the depth of each was not known. Water from the six wells was neither treated nor filtered prior to distribution. The four system B wells and one of two system A wells were permitted and approved by NHDES. The second system A well (well X), was located ∼41 ft (12·5 m) from a brook. NHDES regulation requires well placement at least 50 ft (15 m) from surface water. Well X had been brought online by the previous owner of the water system without a NHDES permit. A site visit to the brook revealed prominent evidence of beaver, including beaver chew in the water and presence of a beaver dam. Upstream from the brook was predominantly national forest with a smaller area occupied by a mixed seasonal community; however, no significant human interaction with the brook was identified.
Water systems in New Hampshire are required to submit monthly water-system distribution samples for coliform testing. Routine samples from the water system collected on 13 August were not contaminated with coliform bacteria. After routine bacteria sampling on 6 September, distribution samples from system A were found to be contaminated with total and faecal coliforms; subsequently, requisite repeat sampling included coliform testing of raw water from the individual wells servicing system A. Faecal coliforms were identified in well X on 10 September, and, as an initial response, well X was hyperchlorinated on 11 September. Repeat coliform testing of distribution samples were negative following hyperchlorination; however, chlorine residuals were high in the samples and invalidated the bacteria assay. After chlorine residuals dropped, distribution samples were again total and faecal coliform-positive indicating ongoing system contamination. On 21 September, well X was disconnected from the system, after which, no faecal coliform-positive distribution samples were obtained.
Laboratory investigation
Human specimens
Human stool specimens from 14 patients were tested at a private laboratory and were FA positive for G. intestinalis. Additional human stool specimens from three patients submitted to NHDHHS and later sent to CDC for subtyping were FA- and PCR-positive for G. intestinalis. The G. intestinalis was identified as assemblage B, and the three specimens exhibited a single subtype that had partial TPI sequence (530 bp) identical to DQ789113 deposited in the GenBank database. The three human specimens were unrelated patients that lived on three different streets in the affected community.
Home water filtration devices
Microscopy testing of the home water filter sample resulted in observation of one suspected Giardia cyst based on particle fluorescence, shape, size, and other morphological characteristics. PCR testing of separate aliquots of the filter sample did not detect the presence of G. intestinalis.
Water distribution samples
Routine water samples collected on 6 September were contaminated with total and faecal coliforms. Daily distribution samples were collected until two consecutive samples collected at least 24 h apart were negative for coliforms, which occurred after the implicated well was removed from the system on 21 September. No water distribution samples were tested for Giardia.
Well water samples
Upon identification of the outbreak, water samples were collected from both wells that served system A. One well, well X (Fig. 1), was contaminated with total and faecal coliforms. Additional water samples from well X were collected in September and tested at a private laboratory; no Giardia cysts were found. Giardia was not detected by microscopy or PCR in final well-water samples collected by NHDES in November and sent to USEPA and CDC for testing. The MPA by USEPA identified diatoms, algae, rotifers, insect/larvae, and other debris in the water and concluded that well X was at moderate risk for being under the direct influence of surface water.
Surface water samples
Numerous Giardia cysts were observed by FA and DIC microscopy analysis of surface water collected from the brook near well X. The microscopy results suggested that the concentration of Giardia cysts in the surface water was ∼40–50 cysts/l. The partial TPI sequences (530 bp) of four positive PCR replicates were identical to each other and to part of the L02116 sequence in the GenBank database, which belongs to the G. intestinalis assemblage B. However, there were five nucleotide differences in the partial TPI gene sequences between the subtype found in three patients and the subtype found in surface water at the positions 746 (T to A), 807 (T to A), 844 (A to G), 1013 (A to C) and 1025 (A to G) of the L02116 sequence, or at the positions 189, 250, 387, 456 and 468 of the 530-bp secondary PCR product.
Beaver samples
Two adult male beavers were collected near a beaver dam in the brook adjacent to well X. Faecal samples collected from the beavers and sent to CDC were PCR-negative for G. intestinalis.
DISCUSSION
We report the largest outbreak of human giardiasis associated with a community drinking-water system in the USA in 10 years. This outbreak affected 63% of responding households in the affected water system, system A; 31 confirmed or probable cases were identified. Illness was significantly associated with consuming tap water. Laboratory and environmental investigations suggest that a groundwater well, well X, was the source of this outbreak. Removal of the implicated well from the water system was followed by cessation of the outbreak.
The well implicated as the source of this outbreak had been brought online without prior regulatory approval by a previous owner and failed to meet the NHDES regulation requiring that wells be placed at least 50 ft (15 m) from surface water [18]. While Giardia was never detected in samples of the well water, laboratory evidence and review of the water system indicated that the unapproved well was faecally contaminated (based on faecal coliform testing) and probably under the influence of surface water contamination (based on MPA testing). Additionally, identification of a suspect Giardia cyst in a home water filter, a proxy for a distribution sample, indicates that Giardia may have been present in the distribution water at some point. Furthermore, epidemiological data showed a strong association between consuming tap water and illness. Unsurprisingly, risk for giardiasis increased with consumption of increasing amounts of tap water, a finding that has been noted in other outbreaks and studies examining the relationship between giardiasis and drinking water [Reference Stuart8, Reference Navin19]. It should be noted that the well-water samples were collected weeks to months after the outbreak and after hyperchlorination of the water system, which may have affected the ability to detect Giardia in the samples.
This outbreak occurred in late August and into early September, which is consistent with known seasonality for giardiasis in the USA. This community was located in a popular tourist area, and several of the homes in the community were only occupied seasonally. As such, at the time of the outbreak, the water system experienced high summer usage. It is our hypothesis that this, coupled with dry weather conditions and the relative close proximity to an adjacent brook, caused the well's cone of depression to expand and intersect surface water, which in turn, was pulled into the well without adequate filtration.
Genotyping and subtyping tools have been used in the characterization of G. intestinalis in sporadic and outbreak cases [Reference Sulaiman14, Reference Sulaiman20, Reference Robertson21]. Subtyping information in conjunction with epidemiological and environmental data can be used to distinguish between sporadic and outbreak-associated cases as well as to assess possible outbreak sources. There are several G. intestinalis assemblages or genotypes, but only assemblages A and B infect humans. These two assemblages have also been found in other mammals, such as domesticated animals, livestock, and beavers [Reference Thompson, Palmer and O'Handley22, Reference Fayer23].
Although most attention regarding the zoonotic potential of G. intestinalis has focused on assemblage A, Sulaiman et al. [Reference Sulaiman14] and Fayer et al. [Reference Fayer23] showed that beavers and muskrats are commonly infected with assemblage B. In the USA, several waterborne outbreaks of giardiasis were previously attributed to contamination of source water by beavers, and giardiasis in backpackers is widely known as ‘beaver fever’ [Reference Navin19, Reference Dykes24, Reference Lopez25]. The G. intestinalis assemblage B subtype found in human cases from this outbreak has substantial sequence differences at the TPI gene (five nucleotide substitutions within the 530-bp PCR target) from the subtype found in a surface water sample taken near the well 1 month after the occurrence of the outbreak. It was also different from other known subtypes in humans but identical to a subtype previously found in beavers in Massachusetts (DQ7899113) [Reference Fayer23] and a Barbary macaque in Italy (GenBank accession no. EU637589). Other TPI sequences in the GenBank differ from those from the outbreak patients by at least five nucleotide substitutions. In contrast, the subtype in the surface water is apparently common, as the 530-bp sequences obtained were identical to AY368169, EF688026, EF688030, EU014503, EU014504, EU014511, EU014515, EU156447, EU272161, and EU272169 in the GenBank, in addition to L02116. This TPI subtype was widely found in humans in the USA, Australia, and Egypt, and ringed seals in Canada [Reference Sulaiman20, Reference Teodorovic, Braverman and Elmendorf26–Reference Foronda28]. Numerous other accession numbers in the GenBank have only one nucleotide substitution, or have identical but shorter TPI DNA sequences.
We could not confirm during the outbreak investigation that beavers were the source of Giardia cysts, because samples from beavers were negative for Giardia, and because only one genetic target (TPI) was analysed. The multilocus analysis was not used because discrepant subtyping results were seen in genetic loci when such an approach was used [Reference Caccio and Ryan29], even in a recent investigation of a large outbreak in Norway, which had over 1500 cases [Reference Robertson21]. In the latter, assemblage B was also found, but analysis of the β-giardin and gdh genes indicated that multiple subtypes were involved. Currently, not enough genotyping work has been done on giardiasis outbreaks to show whether assemblage B causes more outbreaks than assemblage A. In the present study, despite the negative PCR result of samples from beavers, the unique subtype previously found in beavers and the presence of beavers in the brook near the well suggest beavers could have been a potential contamination source. Because giardiasis is a common parasite in a wide variety of species, humans and other domestic and wild animal species cannot be dismissed as possible sources of the outbreak. The finding of a different assemblage B subtype in the surface water 1 month after the outbreak was expected, as the prevalence of Giardia cysts in surface water in the USA and Canada is very high [Reference Keeley and Faulkner30–Reference Wallis33].
There were several limitations to this investigation including (1) low response rate to the survey (probably due to departure of seasonal residents), (2) cases may have been missed if their primary residence was in another state, (3) human and water Giardia subtypes were not identical, and (4) collection of water and beaver samples took place weeks to months after the outbreak. Since Giardia cyst shedding can be sporadic, previous investigations have demonstrated that microscopic examination of an infected beaver's intestinal tissue may reveal Giardia trophozoites despite its stool testing negative [Reference Thompson, Palmer and O'Handley22, Reference Lopez25, Reference Dunlap and Thies34]. Repeated testing is generally recommended, which along with trophozoite analysis was not feasible in this investigation.
Because the outbreak occurred over several weeks and was caused by Giardia, a parasite efficiently transmitted through person-to-person contact, potential secondary cases were included in the statistical analysis, which may have resulted in an underestimation of the association between consuming tap water and illness. The case definition did not include an exclusion for potential secondary cases since investigators were interested in capturing all illness associated with the outbreak. Additionally, exposures were assessed through a household survey but analysed for each individual, which may have decreased the statistical power to detect significant household-level exposures such as use of a home water filtration device. Finally, because of the geographic association of cases and the issuance of the boil water order in the community, the epidemiological investigation focused on drinking-water exposures; no other known sources of giardiasis were assessed.
This outbreak demonstrates the importance of collaboration between health officials, environmental officials, laboratorians, and water system operators, which was critical to determining the outbreak's source. The coordinated, rapid response by investigators prevented additional cases from occurring, which was demonstrated by the fact that cases stopped occurring after the implicated well was disconnected from the water system. Adherence to state and federal drinking-water regulations related to placement of wells and associated treatment is essential and could possibly have prevented this drinking-water-associated outbreak.
ACKNOWLEDGEMENTS
We thank the following persons for their contributions to study design, data collection, and specimen collection and testing: Michael Arrowood, Michael Beach, Michele Hlavsa, Jonathan Yoder, Stephanie Johnston, and Theresa Dearen, Division of Parasitic Diseases, Centers for Disease Control and Prevention; Sarah Krycki, Wendy Lamothe, New Hampshire Department of Health and Human Services; Tylor Young, New Hampshire Department of Safety; Anthony Musante, United States Department of Agriculture, Wildlife Services; Marcel Belaval, Stephanie Harris, United States Environmental Protection Agency. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the NH Department of Health and Human Services, the NH Department of Environmental Services, or the Centers for Disease Control and Prevention. The use of trade names and names of commercial sources is for identification only and does not imply endorsement by these agencies.
DECLARATION OF INTEREST
None.






