INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) infections cause illness ranging from mild diarrhoea to haemorrhagic colitis and life-threatening haemolytic uraemic syndrome (HUS) [Reference Tarr, Gordon and Chandler1]. In the USA, the serogroup most frequently isolated, most strongly associated with HUS, and responsible for the most outbreaks is O157 [Reference Rangel2, Reference Mody3]. Over 50 non-O157 STEC serogroups also cause illness [Reference Strockbine, Kaper and O'Brien4]. Increasing use of Shiga toxin assays by clinical laboratories in recent years has resulted in increased detection of these infections [Reference Hoefer5, Reference Gould6]. In 2010, for the first time, non-O157 STEC infections detected though active sentinel surveillance outnumbered O157 STEC infections [7].
To assess modes of transmission, we describe epidemiological characteristics of all outbreaks of non-O157 STEC infection reported in the USA to 2010. In addition, we assess how outbreak characteristics vary by the presence of STEC virulence factors.
METHODS
Outbreak reports and definitions
An outbreak was defined as epidemiologically linked illnesses (clustered in time or space) with ⩾2 persons having culture-confirmed non-O157 STEC infection of the same serogroup. Single-aetiology outbreaks were defined as isolation of only one serogroup. Multiple-aetiology outbreaks were defined as isolation of only one serogroup of non-O157 STEC from ⩾2 ill persons and laboratory evidence of another enteric pathogen in ⩾2 ill persons. Another enteric pathogen was defined as a pathogen other than non-O157 STEC, or another serogroup of non-O157 STEC, or the same non-O157 STEC serogroup that differed by ⩾4 bands by pulsed-field gel electrophoresis (PFGE) [Reference Tenover8].
Since the 1970s, state and local health departments have voluntarily reported waterborne and foodborne disease outbreaks to designated surveillance systems at the Centers for Disease Control and Prevention (CDC) [Reference Yoder9, Reference Bean10]. Before 2009, states informally reported outbreaks of enteric illness transmitted through contact with animals or ill persons and outbreaks with unknown transmission mode; in 2009, states began reporting these outbreaks through the National Outbreak Reporting System (NORS).
We used four methods to identify outbreaks: (1) queries of the Waterborne Disease Outbreak Surveillance System for outbreaks during 1971–2010, the Foodborne Disease Outbreak Surveillance System (FDOSS) for outbreaks during 1973–2010, and NORS for outbreaks transmitted by contact with animals or persons or by unknown mode during 2009–2010, (2) review of informally reported outbreaks that occurred before 2009, (3) direct contact with all state health departments to ascertain unreported outbreaks, and (4) searches of Medline and health department websites. When discrepancies were found between these sources, we used information from published reports or, if available, updated information from state health departments.
Information collected included setting of exposure, date of first illness, state(s) of exposure, pathogens detected, number of illnesses (laboratory-confirmed and clinically compatible cases), mode of transmission, food vehicle (if applicable), demographic characteristics, and frequency of various symptoms, hospitalization, reported HUS, and death.
We defined outbreak onset as onset date of the first illness. For foodborne outbreaks in which exposures were not limited to a single event or location, setting was classified as community. Most outbreaks were classified into one of four transmission modes: foodborne, waterborne, person-to-person, and animal contact. Outbreaks in childcare centres with no reported mode of transmission were classified as person-to-person. An outbreak was classified as foodborne if a food vehicle was reported, or if there was exposure to a common meal or food establishment. Because outbreak surveillance does not collect laboratory values necessary to confirm HUS, we defined HUS as illnesses reported as HUS by state health departments; it is possible that some of the reported cases did not meet widely accepted HUS case definitions [Reference Tarr and Karpman11].
We defined reported food vehicles as foods implicated through either microbiological or epidemiological evidence and reported to CDC or published in Medline listed reports. If a food contained a single contaminated ingredient or all ingredients belonged to one of 17 defined commodities, the food was classified as that one commodity [Reference Painter12]. Outbreaks that could not be assigned to one commodity, or for which the report contained insufficient information for commodity assignment, were not attributed to any commodity. We defined outbreaks with exposure to the implicated vehicle in more than one state as multi-state.
Isolate characterization
We reviewed CDC records for isolates from outbreaks and identified additional isolates by providing lists to states of isolates characterized at CDC to see if any were from patients in an outbreak. For outbreaks for which CDC had no isolate, we contacted state health department laboratories to collect information on strains characterized there.
CDC's National E. coli Reference Laboratory serotyped non-O157 STEC isolates and tested for virulence genes stx 1 and stx 2 (encode Shiga toxins 1 and 2), eae (encodes intimin), and the putative virulence factor, ehxA (encodes enterohaemolysin), as described previously [Reference Brooks13]. Different methods may have been used at state public health laboratories, but results were accepted irrespective of methodology.
Statistical analysis
We compared mode of transmission of single-aetiology and multiple-aetiology outbreaks and characteristics of single-aetiology outbreaks by mode of transmission and by isolate virulence factors using the χ 2 or Fisher's exact (if an excepted cell size was <5) tests for categorical variables and the Kruskal–Wallis test for continuous variables.
RESULTS
We identified 46 outbreaks involving non-O157 STEC infection that caused 1727 illnesses (477 laboratory-confirmed), 144 hospitalizations, and one death in 26 states (Fig. 1). The first outbreak was reported in 1990 and the number reported increased after 1998. Most (59%) occurred during 2007–2010 (Fig. 2).
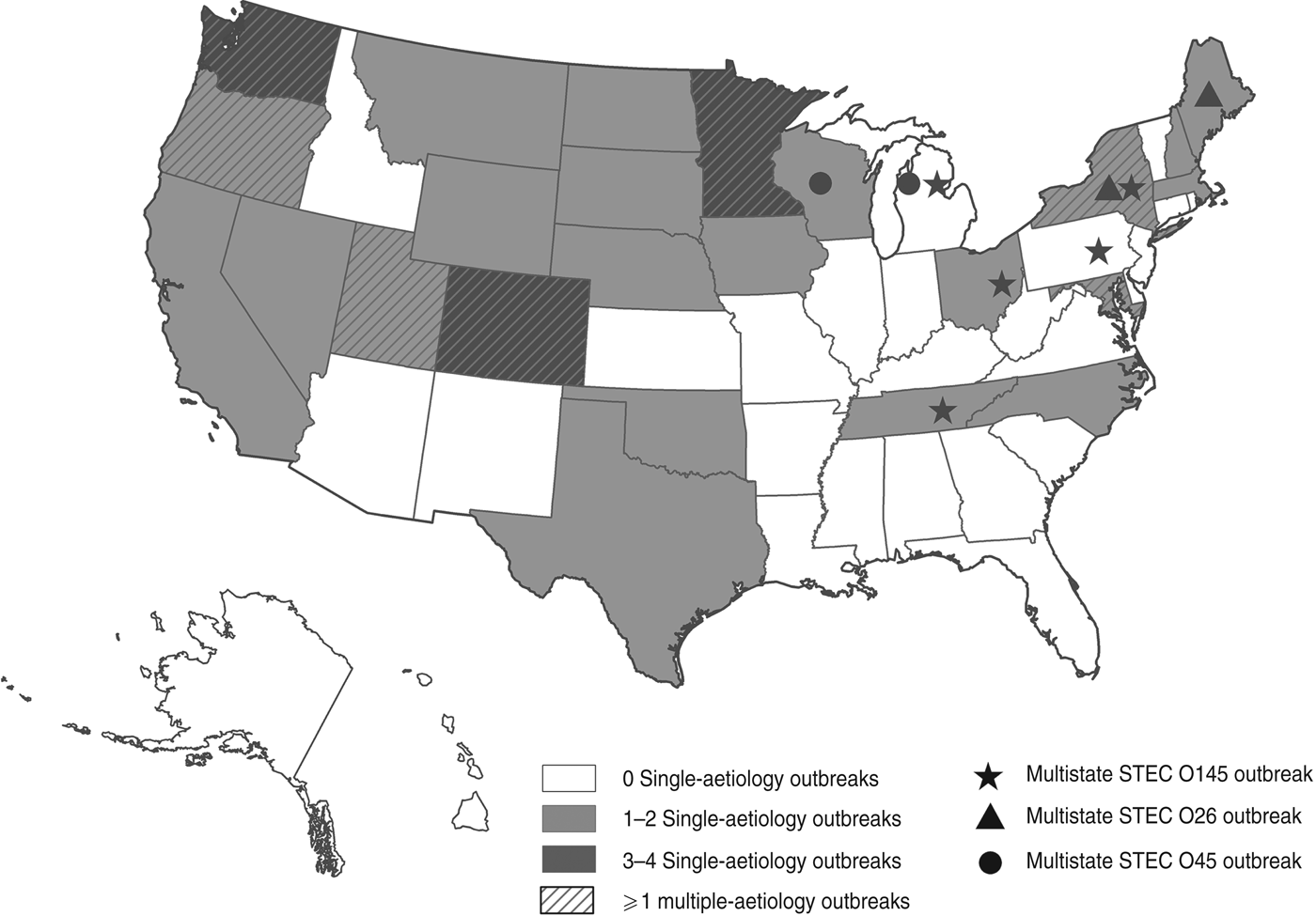
Fig. 1. Outbreaks of non-O157 STEC infection, by state to 2010.
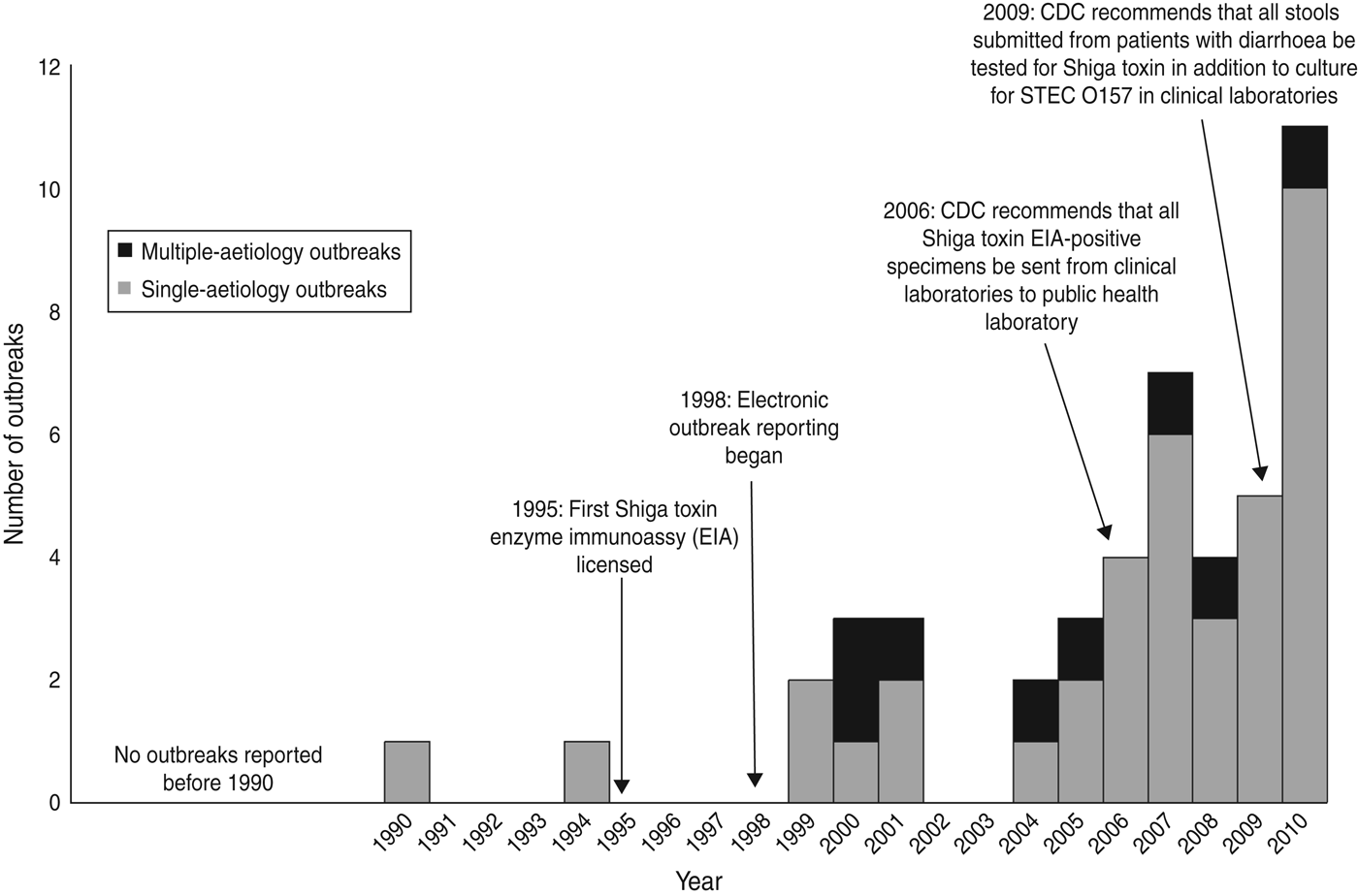
Fig. 2. Number of outbreaks of non-O157 STEC infection, by year to 2010.
Single-aetiology outbreaks
Of 46 outbreaks, 38 (83%) were of single-aetiology, which accounted for 886 (51%) illnesses. Of patients with recorded information, 112 (13%) of 859 were hospitalized, 289 (36%) of 836 reported bloody stool and 39 (4%) of 875 had HUS. One death occurred in a STEC O111 outbreak. Serogroups O111 (14 outbreaks) and O26 (n = 11) accounted for 66% of outbreaks, followed by O45 (n = 4), O103 (n = 2), O121 (n = 2), O145 (n = 2), O104 (n = 1), O165 (n = 1), and O undetermined (n = 1) (Table 1). Most outbreaks (58%) began during June–September. The median outbreak size was 7·5 illnesses (range 2–344). Outbreaks occurred throughout the country (Fig. 1). There were three multistate outbreaks, all in 2010.
Table 1. Single-aetiology outbreak of non-O157 STEC, 1990–2010
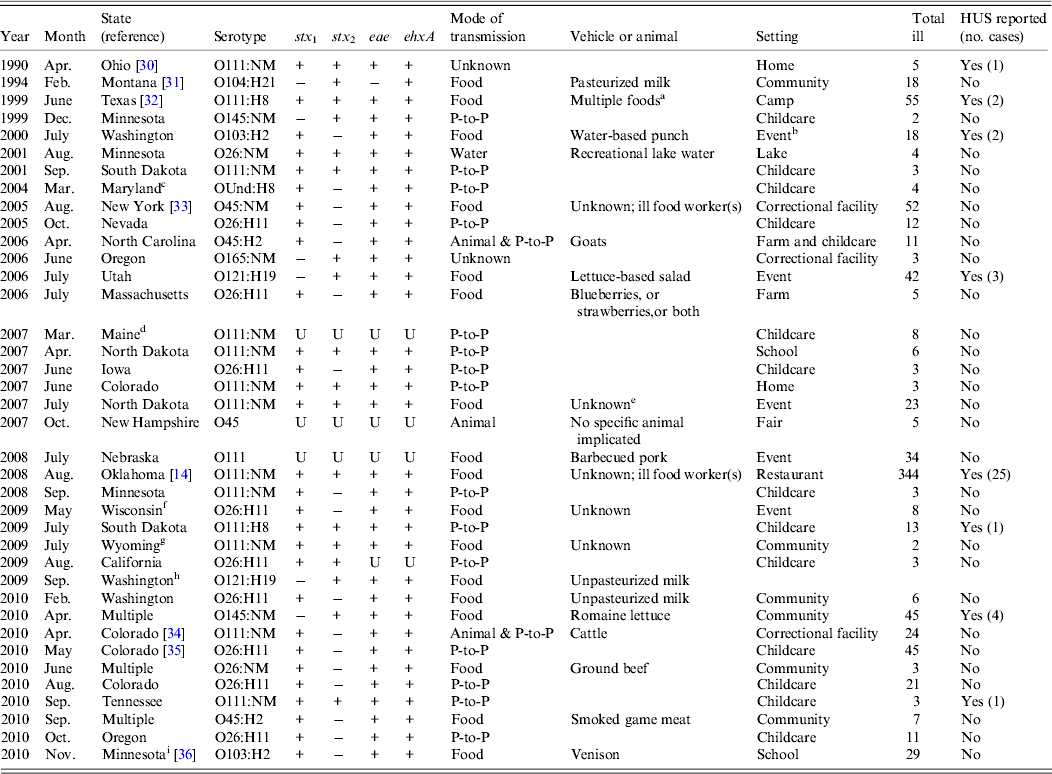
STEC, Shiga toxin-producing Escherichia coli; HUS, haemolytic uraemic syndrome; NM, non-motile; OUnd, O undetermined; P-to-P, person-to-person; U, unknown.
a Cob corn, food from salad bar, dinner roll, and ice.
b Events were banquets, wedding receptions, catered events, customer appreciation days, and family outings.
c Rotavirus was isolated from one patient.
d One patient tested positive for Cryptosporidium.
e The reporting state suspected ground beef as a vehicle on the basis of common food exposures at a catered meal.
f The reporting state suspected ground beef as a vehicle on the basis of reported consumption of undercooked ground beef.
g The reporting state suspected ground beef as a vehicle on the basis of tracebacks of consumed ground beef to the same source.
h E. coli O157:H7 was isolated from one patient.
i Additional patients had E. coli O145:NM isolated; however, neither isolate had Shiga toxin genes. One patient had norovirus identified.
Foodborne
Seventeen (45%) outbreaks were foodborne. The median size was 18 illnesses (range 2–344), which was significantly larger than single-aetiology outbreaks with other modes of transmission (five illnesses, range 2–45, P = 0·02). Foodborne outbreaks accounted for 36 (92%) of all HUS cases.
A food vehicle was reported for 12 outbreaks and 10 could be classified into commodities: dairy (n = 3), leafy vegetables (n = 2), game meat (n = 2), beef (n = 1), pork (n = 1), and fruits or nuts (n = 1). In one of the remaining two outbreaks the vehicle was unclassifiable (water-based punch); in the other, multiple food vehicles including lettuce-containing salads, corn, dinner rolls, and ice, were implicated. Two multistate outbreaks led to food recalls: STEC O145 in Romaine lettuce and STEC O26 in ground beef.
Two of five outbreaks with no identified vehicle, including the largest outbreak of non-O157 STEC infections reported in the USA, reported ill food workers as a possible contributing factor [Reference Bradley14]. Last, in the remaining three outbreaks, ground beef was suspected by states as a possible vehicle on the basis of common food exposures at a catered meal, reported undercooking of ground beef at a barbecue, or tracebacks of ground beef to the same source; however, the states did not feel evidence was sufficient to report ground beef as a vehicle.
Person-to-person
Illnesses in 15 (39%) outbreaks were transmitted from one person to another. The median size was four illnesses (range 2–45). Thirteen (87%) occurred in childcare centres. One outbreak occurred in a home setting and another in an elementary school. Two outbreaks (caused by serotypes O111:non-motile and O111:H8) resulted in two HUS cases.
Other transmission modes
Four outbreaks were attributed to water (n = 1), animal contact (n = 1), or an unknown mode (n = 2). The remaining two outbreaks were propagated through contact with both animals and ill persons. In a STEC O45 outbreak, the index patient visited a goat farm and then attended daycare facilities where other children became ill; other persons who only visited the goats also became ill. In a STEC O111 outbreak in a correctional facility, a subset of ill inmates worked directly with cattle at a dairy and probably spread illness to others.
Multiple-aetiology outbreaks
Eight (17%) of the 46 outbreaks were multiple-aetiology, accounting for 841 (49%) illnesses. Of patients with recorded information, 32 (4%) of 797 were hospitalized and 150 (19%) of 788 reported bloody stool; none developed HUS or died. Two outbreaks were associated with multiple non-O157 STEC strains: one with >1 serogroup and one with a single serogroup displaying two PFGE patterns. The most commonly isolated serogroup was O111 (4 outbreaks), followed by O26 (n = 2), with one outbreak each for O69, O84, O121, O141, O145, and O undetermined. The proportion of patients with culture-confirmed non-O157 STEC infections was generally small relative to those with other laboratory-confirmed infections (Table 2). Other pathogens detected in six outbreaks were Cryptosporidium (n = 3), STEC O157:H7 (n = 3), Campylobacter (n = 3), Shigella (n = 1), Salmonella serotype Typhimurium (n = 1), and norovirus (n = 1). The most frequent pathogen pairings were STEC O111 and Cryptosporidium (n = 3), and STEC O111 and Campylobacter (n = 3). Most (75%) multiple-aetiology outbreaks occurred during June–September.
Table 2. Multiple-aetiology outbreaks in which non-O157 STEC were isolated, USA, 1990–2010
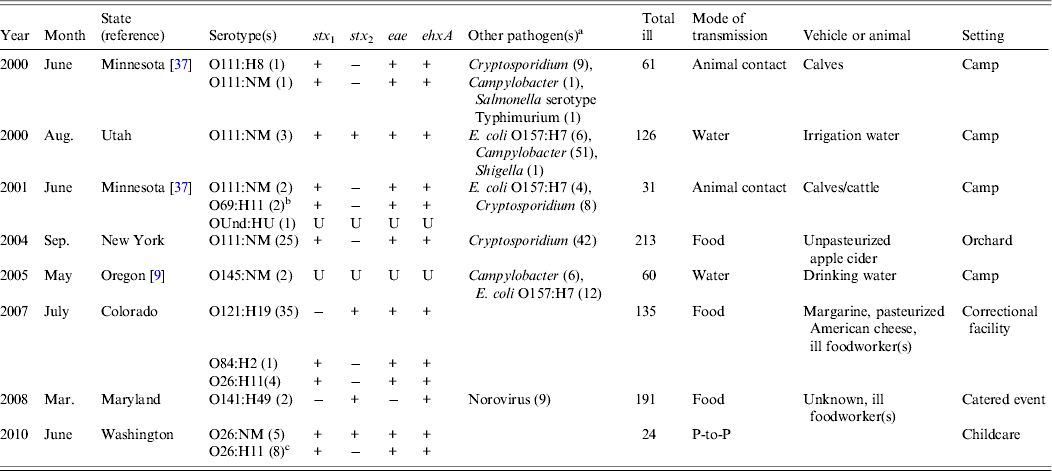
STEC, Shiga toxin-producing Escherichia coli; NM, non-motile; Und, undetermined; U, unknown; P-to-P, person-to-person.
a Number isolated from patients, not mutually exclusive.
b These specimens were originally reported as O rough:H11 in the reference and it was concluded that they were O51:H11 for which the O51 antigen could not be identified. These specimens were retested at CDC in 2011 and were determined to be O69:H11.
c PFGE patterns differed by four bands.
Three multiple-aetiology outbreaks were associated with food; the vehicle in one was unpasteurized apple cider. In the two remaining outbreaks ill food workers were reported as a possible contributing factor: one additionally implicated two dairy products.
The other modes of transmission in multiple-aetiology outbreaks were animal contact (n = 2), water (n = 2), and person-to-person (n = 1). Both outbreaks transmitted through animal contact or water occurred in camp settings.
Compared with single-aetiology outbreaks, multiple-aetiology outbreaks had more illnesses transmitted though water or animal contact, and more illnesses were seen in children aged 5–19 years (Table 3).
Table 3. Transmission, demographic, and clinical characteristics of patients in single- and multiple-aetiology outbreaks a
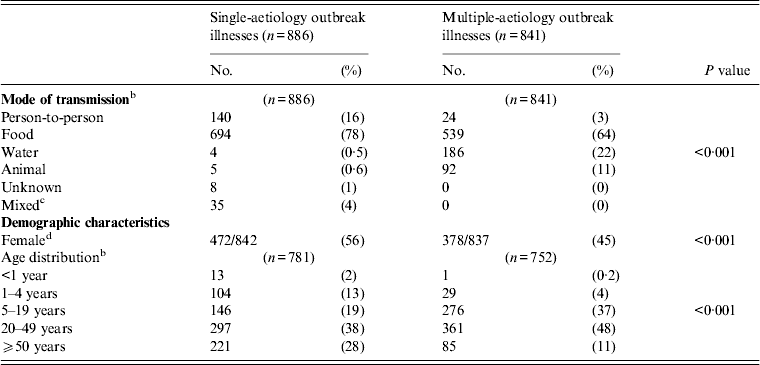
a All P values based on the χ 2 test unless otherwise noted.
b Fisher's exact test.
c Mixed modes of transmission were defined as outbreaks with ⩾1 mode of transmission. Two outbreaks met this criterion. Both involved contact with animals and person-to-person contact.
d Gender differences ceased to be significant when four outbreaks of ⩾50 illnesses in which ⩾85% of patients are of one gender were excluded: a STEC O111 outbreak at a cheerleading camp (55 illnesses, 96% female), a STEC O45 outbreak at a correctional facility (52 illnesses, 100% male), an outbreak involving STEC O121:H19, O26:H11, and O84:H2 infections at a correctional facility (135 illnesses, 85% male), and an outbreak involving STEC O111, Campylobacter, Shigella, and STEC O157:H7 at a football camp (126 illnesses, 94% male). Removing these outbreaks increases the percentage of female patients to 57% and 61% in single-aetiology and multiple-aetiology outbreaks, respectively (P = 0·14).
Virulence factor characterization
Shiga toxin information was available for 35/38 single-aetiology outbreak strains and for 11 strains from 7/8 multiple-aetiology outbreaks (Tables 1 and 2). All but one strain in each group (single-aetiology and multiple-aetiology outbreaks), for which information was available, was eae positive and all were ehxA positive.
Outbreaks with stx 2-positive strains (with or without stx 1) were significantly more likely to have HUS cases reported than outbreaks with stx 1-only positive strains (32% vs. 4% of outbreaks, P = 0·02). In an analysis of single-aetiology outbreaks, 7% of patients in outbreaks with a stx 2-positive strain (with or without stx 1) developed HUS compared with 0·8% of patients in outbreaks with a stx 1-only positive strain (P< 0·001).
DISCUSSION
Non-O157 STEC strains are increasingly recognized as causes of enteric disease outbreaks in the USA. Of the 38 outbreaks we identified, none occurred before 1990 and over half occurred during 2007–2010. Most outbreaks were transmitted through contaminated food or from one person to another. Nearly one-fifth of all outbreaks involved more than one pathogen, and these multiple-aetiology outbreaks were epidemiologically different from single-aetiology outbreaks. Over half of all outbreaks involved STEC serogroups O111 or O26.
The increasing detection of non-O157 STEC outbreaks coincides with a marked rise in reporting of sporadic non-O157 STEC infections, which is related to the increasing use of enzyme immunoassays or polymerase chain reaction by clinical laboratories to detect Shiga toxin (or the genes that encode them) in the stool of patients with diarrhoea [Reference Hoefer5, Reference Gould6]. These tests, first licensed in 1995, are currently the only widely available, practical means of detecting non-O157 STEC in clinical specimens.
Although non-O157 STEC are estimated to cause nearly twice as many infections as STEC O157 in the USA [Reference Scallan15] the number of detected outbreaks caused by non-O157 STEC is substantially less. This is probably due in part to more severe illness in persons with STEC O157 than in the broad group of non-O157 STEC infections [Reference Gould6]. Moreover, clinical laboratories can more readily identify STEC O157. Unlike for STEC O157, few states were routinely subtyping non-O157 STEC isolates by PFGE until the past 5 years; submitting PFGE patterns of subtyped isolates to PulseNet facilitates detection of outbreaks. As more clinical laboratories test for Shiga toxin and more non-O157 STEC infections are reported, we can expect greater detection of non-O157 STEC outbreaks.
The number of outbreaks we describe does not include all those reported by state health departments. Our case definition (at least two epidemiologically linked culture-confirmed cases) excluded seven outbreaks [16]. In one, non-O157 STEC were isolated from two persons but the serogroups differed. In the other six, only one case was culture-confirmed (four with STEC O121, two with O111). Three included reports of HUS, including one published as an O121 outbreak [Reference McCarthy17].
The proportion of single-aetiology non-O157 STEC outbreaks attributed to food (43%) and, especially, person-to-person transmission (35%) differs from that found for US STEC O157 outbreaks during 1982–2002 (52% foodborne, 14% person-to-person) [Reference Rangel2]. These differences may relate to past challenges in detecting non-O157 STEC infections and, consequently, limited detection of outbreaks via laboratory-based surveillance. A greater portion of non-O157 STEC outbreaks may have been detected because patients or physicians contacted health departments after noticing a localized cluster of illness. Our findings suggest this may be especially true for outbreaks in childcare centres, the setting for nearly 90% of outbreaks due to person-to-person transmission, and camps, the setting of half of multiple-aetiology outbreaks. Person-to-person transmission also accounts for a considerable number of the non-O157 STEC outbreaks reported from other countries [Reference Boudailliez18, Reference Sonoda19]. Likewise, the historical effect of limited laboratory surveillance of non-O157 STEC would probably result in disproportionately underdetecting foodborne outbreaks. Without sensitive surveillance, contaminated commercially distributed products that cause widely dispersed infections may go unnoticed. All three multistate outbreaks of non-O157 STEC infection were foodborne and detected through the combination of appropriate testing in clinical laboratories and PulseNet surveillance.
Of the single-aetiology foodborne disease outbreaks, vehicles included those implicated in STEC O157 outbreaks in the USA, including leafy vegetables, dairy, game meat, and ground beef [Reference Rangel2]. Of reported non-O157 STEC foodborne outbreaks in other countries, implicated foods have also included beef and dairy products [Reference De20–Reference Ethelberg23]. We are unaware of any reports from outside the USA that implicate leafy vegetables as a non-O157 STEC outbreak vehicle. However, contaminated raw sprouts caused a severe STEC O104:H4 outbreak in Europe [Reference Buchholz24]. The first reported non-O157 STEC outbreak in the USA associated with raw sprouts occurred in 2012 [25].
Environmental exposures play an important role in multiple-aetiology outbreaks. Exposures to cattle or contaminated water (for drinking, swimming, or irrigation) accounted for a greater proportion of illnesses in multiple-aetiology than in single-aetiology outbreaks. Half of all multiple-aetiology outbreaks occurred at camps, where children may have contact with animals and untreated water. This may explain the greater frequency of illnesses in children aged 5–19 years in multiple-aetiology outbreaks compared with single-aetiology outbreaks. Furthermore, the other pathogens most frequently identified in these outbreaks (Cryptosporidium, E. coli O157, Campylobacter) are carried by ruminant animals and have caused waterborne outbreaks. Similarly, animal contact may have caused multiple-aetiology outbreaks in Belgium and Australia [Reference De20, Reference Hanna26].
Outbreaks of infection with STEC strains that contain stx 2 or eae genes carry a greater risk of bloody diarrhoea or HUS [Reference Ethelberg27] probably making them easier to detect. Enterohaemolysin (encoded by ehxA) is a putative virulence factor. A greater percentage of the 34 single-aetiology outbreak strains we identified carried stx 2 (50%), eae (97%), or ehxA (100%) than commonly seen in non-O157 STEC isolated from ill persons. In a USA convenience sample of >900 non-O157 STEC isolates from ill persons, 39% carried stx 2, 84% carried eae and 86% carried ehxA [Reference Brooks13].
The clinical characteristics of infections caused by STEC of different serogroups may affect the number of outbreaks observed. Two serogroups were responsible for two-thirds of the 38 single-aetiology outbreaks, O111 (38%) and O26 (29%). The high frequency of STEC O26 outbreaks could simply reflect the high frequency of STEC O26 infections in the USA. From 2000 to 2010, 1708 non-O157 STEC infections with a known serogroup were reported to FoodNet; O26 accounted for 26%, followed by O103 (22%) and O111 (19%) [Reference Gould6]. Alternatively, O111 and O26 may predominate because of low infectious doses, facilitating person-to-person transmission [Reference Paton28]. Nearly 90% of person-to-person outbreaks were caused by these two serogroups. The high frequency of STEC O111 outbreaks may relate to the greater propensity of this serogroup to produce Stx2 and cause severe illness compared with STEC O26 and STEC O103, thereby facilitating outbreak detection [Reference Mody3, Reference Brooks13]. Serogroup O111 caused 83% of all outbreaks with HUS cases.
The percentage of illnesses reported as being complicated by HUS in single-aetiology outbreaks was relatively high (4%). Only about 1% of non-O157 STEC infections in the USA lead to physician-diagnosed HUS compared with 11% of STEC O157 infections [Reference Gould6]. This higher than expected percentage of reported HUS in non-O157 STEC outbreaks suggests that outbreaks with more severe illnesses were more likely to be detected. Alternatively, reporting officials may not have applied appropriate criteria in reporting HUS cases. We were unable to verify that all reported HUS cases met widely accepted laboratory-defined HUS definitions because HUS-defining laboratory values including serum creatinine, haemoglobin (or haematocrit), and platelet count are not collected through outbreak surveillance. However, many of the reported cases of HUS (25/39, 64%) in our analysis were from a single outbreak of STEC O111 infections that did apply laboratory criteria to define HUS [Reference Bradley14].
Reports to the outbreak surveillance system include both laboratory-confirmed and other epidemiologically linked illnesses [16]. In this analysis, the percentage of reported infections that were not laboratory confirmed was relatively large (72%). Thus, for every laboratory-confirmed illness reported (28% of all illnesses) in the outbreaks described there were 3·6 non-confirmed illness reported. It is possible that some illnesses were misclassified as being part of an outbreak. However, it is unlikely that inclusion of some non-confirmed cases would have resulted in considerable overestimation of outbreak size. Most people with diarrhoeal illness do not seek medical care and even fewer submit a stool specimen. It has been estimated that for every confirmed non-O157 STEC infection there may be as many as 107 undetected cases in the population [Reference Scallan15].
Because of increasing use of assays that detect Shiga toxins in clinical laboratories, non-O157 STEC strains have increasingly been detected as causes of outbreaks in the USA during the past two decades. These infections are acquired through several routes of transmission. Stools of patients with community-acquired diarrhoea for which an aetiology is sought should be tested for Shiga toxin and all positive samples should be sent to public health laboratories for STEC isolation and characterization [Reference Gould29]. Such an approach can identify all pathogenic non-O157 STEC including important novel strains such STEC O104:H4 and can improve our understanding of these important pathogens.
ACKNOWLEDGEMENTS
We thank all of the public health practitioners in state and local health departments, without whom this report would not have been possible. We are grateful to Ms. Katherine Heiman for her assistance with preparing figures.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
DECLARATION OF INTEREST
None.








