Introduction
Inbreeding decreases the performance of dairy cattle by directly or indirectly affecting various productive and reproductive parameters (Gutiérrez-Reinoso et al., Reference Gutiérrez-Reinoso, Aponte, Cabezas, Rodriguez-Alvarez and Garcia-Herreros2020). During the 1970s, in countries such as the United States, the degree of inbreeding was recorded to be in the order of 3–6% in the Holstein breed (Young, Reference Young1984). This problem occurs because breeding programs in dairy cattle incentivize and reinforce matings between genetically related individuals, driving the phenomenon called inbreeding depression (Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019). Even though estimates of genomic values have received much attention in recent years, estimates of inbreeding depression in dairy cattle are rare in the literature (Maltecca et al., Reference Maltecca, Tiezzi, Cole and Baes2020). Several authors suggest a variable influence of genomic selection on the effect of inbreeding (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017; Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019). In dairy cattle, the impact of inbreeding depression in recent generations is projected to drive very adverse effects compared to old inbreeding (distant generations) (Makanjuola et al., Reference Makanjuola, Maltecca, Miglior, Schenkel and Baes2020). However, inbreeding may exert a detrimental effect only on specific traits (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019; Gutierrez-Reinoso et al., Reference Gutierrez-Reinoso, Aponte and Garcia-Herreros2021).
Implementing genomic evaluations has drastically changed how breeding systems are conducted in dairy cattle breeds (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017). Traditionally, pedigree-derived data was highly valuable for assessing the genetic diversity of non-genotyped individuals (Sonesson et al., Reference Sonesson, Woolliams and Meuwissen2012). However, genomics has enabled more precise studies of the proportion of the inbreeding patterns across the genome (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017; Sell-Kubiak et al., Reference Sell-Kubiak, Czarniecki and Strabel2018). Thus, genomics-based estimates started to be used to manage genetic diversity in animal selection programs, including dairy cattle (Sonesson et al., Reference Sonesson, Woolliams and Meuwissen2012).
Inbreeding has been described as a cause of the reduced performance of productive and reproductive traits in dairy cattle by increasing the frequency of deleterious or non-deleterious recessive homozygous genotypes leading to the loss of genetic dominance and other non-additive effects (VanRaden, Reference VanRaden1992; McParland et al., Reference McParland, Kearney and Berry2009; Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014; Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015; Gutiérrez-Reinoso et al., Reference Gutiérrez-Reinoso, Aponte, Cabezas, Rodriguez-Alvarez and Garcia-Herreros2020). Consequently, inbreeding depression can be reduced by minimizing overall inbreeding and avoiding the production of recessive homozygous offspring (Leroy, Reference Leroy2014; Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014).
The accumulation of annual genetic gain has gathered momentum for major dairy cattle breeds (Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019) due to the implementation of genomic selection during the last decades. Still, unfortunately, the annual loss of genetic diversity has increased considerably, reflecting the emergence of recent inbreeding (Makanjuola et al., Reference Makanjuola, Maltecca, Miglior, Schenkel and Baes2020). Therefore, understanding the impact of genomic selection on genetic diversity and the factors involved in different genetic traits (productive, reproductive, type, health, conformation, and adaptation, among others) as well as inbreeding rates based on genomics and pedigree estimations is critical for the development of dairy cattle selection programs (Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014; Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019; Gutierrez-Reinoso et al., Reference Gutierrez-Reinoso, Aponte and Garcia-Herreros2021).
Despite the evident negative influence of inbreeding on several genetic traits in dairy cattle (McParland et al., Reference McParland, Kearney and Berry2009; Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014; Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015; Gutiérrez-Reinoso et al., Reference Gutiérrez-Reinoso, Aponte, Cabezas, Rodriguez-Alvarez and Garcia-Herreros2020), the mating of genetically related individuals continues to be carried out (Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019). Breeding organizations continue to develop breeding programs involving very close or genetically related genetic lines, intending to fix different traits of interest (Gandini et al., Reference Gandini, Stella, del Corvo and Jansen2014). This issue has led to a vicious circle, negatively impacting genomic and phenotypic traits related to production, reproduction, conformation, health, and adaptability. For example, Holstein bulls show a higher annual genetic variability loss than other dairy breeds (Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019). To progressively generate understanding in the industry for the need to mitigate the negative inbreeding derived effects in dairy cattle breeds, urgent characterization, and dissemination of information on the extent of inbreeding and inbreeding coefficients of specific bulls are becoming extremely important.
The present review sets out the primary objectives of providing a broader understanding of the importance of genomic analysis and exploring possible applications to mitigate the undesirable effects of inbreeding depression on different traits of production, reproduction, health and welfare, linear conformation and adaptability. We will address topics related to the use and influence of genomic analysis on genotypic traits affected by inbreeding in dairy cattle. This review paper is designed and organized as follows: a brief history of the emergence of the concept of inbreeding/endogamic depression, the impact of the negative effect of inbreeding on production traits, reproduction, animal health-welfare, linear conformation, and adaptability indices in different cattle operation systems, as well as causal and highly predictive genetic variants potentially key to the prediction of other complex traits. Furthermore, we will provide evidence that the control of inbreeding depression should focus on current methodologies applied to genomic selection. Promising new approaches to dairy cattle genomic selection based on genomic analysis show excellent potential to control the genetic gain of genomic traits that influence different aspects of dairy cattle. These tools are essential to improve the prediction of the effects derived from increased homozygosity. However, considerable challenges exist to integrate a genomic analysis model that increases the accuracy of prediction for different genetic traits while at the same time detecting a greater or lesser increase in inbreeding depression. Therefore, monitoring the degree of homozygosity in dairy cattle becomes a critical factor, instrumental for controlling and determining the possible adverse effects derived from it. Consequently, follow-up of the phenomenon of homozygosity must be considered to counteract endogamic effects. This way, we can ensure a sustainable future and profitability during the productive life of dairy cattle across generations.
Inbreeding depression: history, background and current status
Inbreeding is the probability of two alleles in an individual being identical by descent due to mating-related individuals. The inbreeding rate is a function of the characteristics of the foundation stock as well as limited population sizes in subsequent generations (Possingham et al., Reference Possingham, McCarthy and Lindenmayer2013). Inbreeding resulting from the mating between animals of standard genetic lines can be associated with a decrease or loss of the biological capacity called inbreeding depression. This concept is advanced by different authors who maintain that high levels of endogamy are undesirable because they diminish genetic diversity and yields of the future generations due to positive or negative influences over different traits (Miglior et al., Reference Miglior, Van Doormaal, Kistemaker, Canada, Network and Swiss2001; Croquet et al., Reference Croquet, Mayeres, Gillo, Vanderick and Gengler2006; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019; Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019) (Fig. 1). The first studies to demonstrate the effects of inbreeding depression were conducted in plants and, later, the concept was extrapolated to animal species.
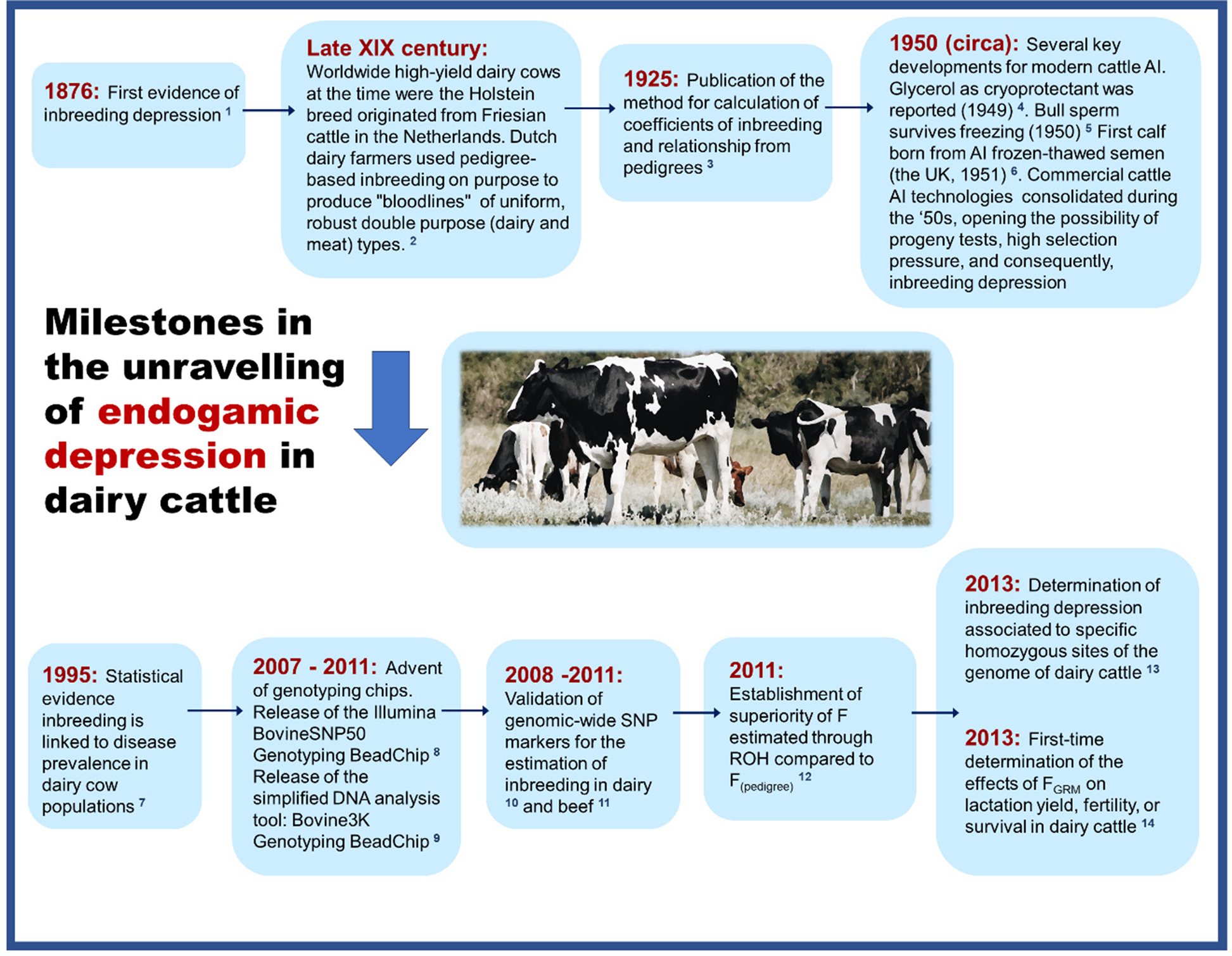
Fig. 1. Milestones in the unraveling of endogamic depression in dairy cattle. AI = artificial insemination; F (pedigree) = coefficient of inbreeding estimated through pedigree; ROH = Runs of homozygosity; F GRM = Inbreeding coefficient based on the genomic relationship matrix. References: (1) (Darwin, Reference Darwi1876); (2) (Theunissen, Reference Theunissen2008); (3) (Wright and McPhee, Reference Wright and McPhee1925); (4) (Polge and Rowson, Reference Polge and Rowson1952); (5) (Bailey, Reference Bailey2017); (6) (Leibo et al., Reference Leibo, Semple and Kroetsch1994); (7) Miglior et al., Reference Miglior, Burnside and Dekkers1995); (8) (BovineSNP50 DNA Analysis Kit, 2021); (9) (Data Sheet: DNA Analysis, 2021); (10) (VanRaden et al., Reference VanRaden, Olson, Wiggan, Cole and Tooker2011); (11) (Hayes and Goddard, Reference Hayes and Goddard2008); (12) (Keller et al., Reference Keller, Visscher and Goddard2011); (13) (Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014); (14) (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013).
Initially, evaluations of the degree and consequences of inbreeding in dairy cattle breeds were based on estimates of pedigree data. Consequently, predictions of inbreeding coefficients used to be fluctuating and imprecise (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017). Today, computerized animal selection programs have substantially improved the estimation of traits by reasonably limiting the degree of inbreeding in subsequent generations (Weigel and Lin, Reference Weigel and Lin2000). In parallel to these studies, a large number of meta-analyses have been conducted on various livestock species, demonstrating that reproductive traits are more severely depressed than other traits (McParland et al., Reference McParland, Kearney, Rath and Berry2007; Ma et al., Reference Ma, Cole, Da and VanRaden2019). Overall, for every 1% increase in the degree of inbreeding, an average decrease of 0.137% of some traits is estimated (Leroy, Reference Leroy2014), including production losses (McParland et al., Reference McParland, Kearney, Rath and Berry2007; Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019). In other cases, inbreeding depression has been observed to cause yield losses for production traits like for instance −32 to −41 kg of 305ME milk, −1.4 to −1.7 kg of 305ME fat, and −1.1 to −1.3 kg of 305ME protein by percent inbreeding (Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015), or reducing fat and protein concentrations by 0.05% and 0.01%, respectively while somatic cell scores increased by 0.03–0.86 units (McParland et al., Reference McParland, Kearney, Rath and Berry2007; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019). (Table 1). Also, due to the effect of inbreeding, 2% more dystocia, 1% more stillbirths, 0.7% more male calves, an increase of calving interval of 8.8 d and increased age at first parturition of 2.5 d have been reported (McParland et al., Reference McParland, Kearney, Rath and Berry2007; Ma et al., Reference Ma, Cole, Da and VanRaden2019) (Table 2).
Table 1. Regression coefficients of inbreeding depression for production and conformation for production traits per 1% increase in inbreeding in dairy cattle (genomics vs. pedigree)

Estimates were readjusted as standardized linear regression coefficients.
Table 2. Regression coefficients of inbreeding depression for reproduction and conformation for ease of calving traits per 1% increase in inbreeding in dairy cattle (genomics vs. pedigree)

AFS_H: Age at first service for heifers, NS_H: Number of services for heifers, NRR_H: 56-day non-return rate for heifers, FSTC_H: First service to conception for heifers, CTFS_C: Conception to the first service for cows, NS_C: Number of service for cows, NRR_C: 56-d non-return rate for cows, FSTC_C: First service to conception for cows. Estimates were readjusted as standardized linear regression coefficients.
Despite the findings of several studies that suggest inbreeding negatively affects productive and other traits (Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010; Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015; Howard et al., Reference Howard, Pryce, Baes and Maltecca2017; Martikainen et al., Reference Martikainen, Tyrisevä, Matilainen, Pösö and Uimari2017; Yurchenko et al., Reference Yurchenko, Daetwyler, Yudin, Schnabel, vander Jagt, Soloshenko, Lhasaranov, Popov, Taylor and Larkin2018; Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019), other authors indicate that inbreeding is unlikely to cause large losses (McParland et al., Reference McParland, Kearney, Rath and Berry2007). Although overdominance and epistasis may contribute to inbreeding depression in dairy cattle, partial dominance is expected to account for the more significant proportion of inbreeding depression. Thus, the degree and timing of inbreeding do not always seem to be negative, considering that recent inbreeding is more detrimental than old inbreeding (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019). In dairy breeds such as Holstein and Jersey, these effects would be accompanied by an increased frequency of unfavorable homozygous recessive genotypes (Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014). Therefore, the estimation of genomic homozygosity seems to be a more accurate tool than the pedigree-based inbreeding coefficient (Kardos et al., Reference Kardos, Luikart and Allendorf2015; Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019).
There is a consensus that the effect of inbreeding at undesirable levels seriously affecting genetic variability, as well as the performance in generations of dairy cattle (Croquet et al., Reference Croquet, Mayeres, Gillo, Vanderick and Gengler2006; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019; Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019; Miglior et al., Reference Miglior, Van Doormaal, Kistemaker, Canada, Network and Swiss2001) is associated to specific genomic regions (Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014). This implies that inbreeding coefficients vary and are inaccurate when their predictions are based solely on pedigree data (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017). One summarizing remark from the above studies is that the continuous use of genomic evaluation as a routine method in bovine genetic improvement programs is highly relevant to positively impact the reduction of the degree of inbreeding in the future generations (de Oliveira Seno et al., Reference de Oliveira Seno, Guidolin, Aspilcueta-Borquis, do Nascimento, da Silva, de Oliveira and Munari2018). However, estimation of genomic homozygosity appears to be a more accurate tool (Kardos et al., Reference Kardos, Luikart and Allendorf2015; Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019), so linking these two factors (genomics and pedigree) significantly improves the estimation of prediction for the most desirable traits of interest in dairy cattle (Weigel and Lin, Reference Weigel and Lin2000).
Inbreeding depression in dairy cattle: production, reproduction, health and welfare, linear conformation and adaptability
Impact of inbreeding depression on production
One of the main advantages of bovine genome sequencing is that it has contributed significantly to the analysis and interpretation of the functionality of genes. This advance leads to opportunities to answer the problem of the overuse of existing genetic lines in dairy breeds that has generated an increase in the consanguinity index (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019). The reported incidence of inbreeding and, consequently, inbreeding depression is evident in several traits, particularly in production traits (Pryce et al., Reference Pryce, Haile-Mariam, Goddard and Hayes2014). Thus, inbreeding generates a substantial effect on milk, fat, and protein derived-traits, and an alteration of the somatic cell count as evaluated through genealogical information of sires and dams of different dairy breeds (Croquet et al., Reference Croquet, Mayeres, Gillon, Hammami, Soyeurt, Vanderick and Gengler2007; Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010; Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013; Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015). In another study involving Jersey animals in which different degrees of inbreeding were estimated over time and their relationship with production and genealogical data was quantified, a greater negative presence of inbreeding in animals of early ages and at the beginning of lactation was found (Thompson et al., Reference Thompson, Everett and Wolfe2000). Also, in other breeds such as Ayrshire, this time introducing genomic analysis, harmful homozygous recessive alleles have been identified to be responsible for the increase in inbreeding depression affecting production traits in general (Martikainen et al., Reference Martikainen, Koivula and Uimari2020). When identical haplotypes are inherited from both parents, inbreeding seems to be related to continuous lengths of homozygous genotypes (runs of homozygosity, ROH). These may be formed with deleterious (lethal) recessive alleles and could be associated with inbreeding depression and consequently with a decreased phenotypic performance. In the Ayrshire dairy breed, several ROHs have been reported to adversely affect production and reproductive traits (Martikainen et al., Reference Martikainen, Koivula and Uimari2020). However, not all ROHs in a region may have negative effects on the trait of interest. Therefore, more efficient control of inbreeding depression could minimize the occurrence of unfavorable haplotypes as homozygous status in breeding programs.
According to Doekes et al. (Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019), inbreeding in Dutch Holstein-Friesian cows decreases animal performance traits (inbreeding depression), but inbreeding may not always be detrimental. In the Holstein breed, the effect of the degree of ancestral and current inbreeding ratifies the harmful effects of inbreeding on production traits (McParland et al., Reference McParland, Kearney and Berry2009). Over time the frequency of deleterious (lethal) alleles decreases due to a selection process called genetic purging, and recent inbreeding may be more harmful than old inbreeding. In both production and reproductive traits, long and short ROHs contributed to inbreeding depression (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019).
Genomics has recently emerged as a tool for the evaluation of inbreeding depression. Several genomic studies have detected the continuous increase of inbreeding indexes associated with deleterious homozygous recessive alleles, responsible for the rise of inbreeding depression affecting production traits in several dairy breeds (Croquet et al., Reference Croquet, Mayeres, Gillon, Hammami, Soyeurt, Vanderick and Gengler2007; Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010; Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013; Dezetter et al., Reference Dezetter, Leclerc, Mattalia, Barbat, Boichard and Ducrocq2015; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019), such as the Ayrshire (Martikainen et al., Reference Martikainen, Koivula and Uimari2020), Jersey (Thompson et al., Reference Thompson, Everett and Wolfe2000) and Holstein (McParland et al., Reference McParland, Kearney and Berry2009). Therefore, the implementation of genomic analysis is essential to understand the functionality and potential of some genes to influence productive traits and establish the size of the association of conventional traits regarding new traits included in genetic improvement programs in recent years.
Effects of inbreeding depression on reproduction
Increased inbreeding rates have been associated with reduced reproductive capacity in major dairy breeds (Martikainen et al., Reference Martikainen, Koivula and Uimari2020). In the Holstein breed, it has been shown that classic, ancestral and future inbreeding has detrimental effects on fertility and survival traits (McParland et al., Reference McParland, Kearney and Berry2009). For decades, the analysis to determine the degree of inbreeding has been performed relying on genealogical data, and adverse effects on reproduction have been reported (Hermas et al., Reference Hermas, Young and Rust1987). For example, pedigree-based estimations of the inbreeding coefficients in the Ayrshire breed show important differences derived from inbreeding depression on the fertility indexes, the prediction through genomic analysis being more precise (Martikainen et al., Reference Martikainen, Tyrisevä, Matilainen, Pösö and Uimari2017). Moreover, studies using solely genealogical data show that the estimation of the inbreeding coefficient had no significant effects on traits such as birth weight and calving ease (McParland et al., Reference McParland, Kearney, Rath and Berry2007). In contrast, a study in adult Holstein cattle showed that inbreeding, assessed by pedigree data, affected the calving interval trait and detected an increased incidence of difficult calvings (Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010). Evaluations of inbreeding depression from genomic data have shown a more accurate and significant prediction of reproductive traits (Martikainen et al., Reference Martikainen, Sironen and Uimari2018) such as decreased oocyte and embryo competition (Perez et al., Reference Perez, Balieiro, Ventura, Bruneli and Peixoto2017), fetal death (Hinrichs and Thaller, Reference Hinrichs and Thaller2011), udder health traits and overall fertility rates (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019) as well as days open and calving ease (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013).
Genomic regions have also been identified that harbor harmful recessive mutations associated with decreased calving rates (Fritz et al., Reference Fritz, Capitan, Djari, Rodriguez, Barbat, Baur, Grohs, Weiss, Boussaha, Esquerré, Klopp, Rocha and Boichard2013) and lethal recessive mutations associated with embryonic death (VanRaden and Miller, Reference VanRaden and Miller2006). Hence, the effects of inbreeding rates on different reproductive traits should be considered in genetic improvement programs in dairy cattle (Martikainen et al., Reference Martikainen, Tyrisevä, Matilainen, Pösö and Uimari2017). For such evaluation, genomic evaluation techniques should be prioritized due to their predictive capacity and greater precision. Although the results of inbreeding estimation using pedigree data are similar to those estimated by genomics for some reproductive traits, certainly genomic estimations are more efficient and accurate (Weller et al., Reference Weller, Ezra and Ron2017). Some authors even claim that there is now evidence of increased predictive accuracy using genomic analyses in dairy cattle breeds in this respect (Martikainen et al., Reference Martikainen, Sironen and Uimari2018, Reference Martikainen, Koivula and Uimari2020). Through genomic methods, the different regions in the genome that have been identified as harboring harmful recessive mutations associated with negative phenotypic effects involve calving rates (Fritz et al., Reference Fritz, Capitan, Djari, Rodriguez, Barbat, Baur, Grohs, Weiss, Boussaha, Esquerré, Klopp, Rocha and Boichard2013), embryonic death (VanRaden and Miller, Reference VanRaden and Miller2006), fertility (Martikainen et al., Reference Martikainen, Tyrisevä, Matilainen, Pösö and Uimari2017), calving interval, dystocic parturition (Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010; Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013), decreased oocyte/embryo competence (Perez et al., Reference Perez, Balieiro, Ventura, Bruneli and Peixoto2017), early fetal death (Hinrichs and Thaller, Reference Hinrichs and Thaller2011), lower fertility rates (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019), and increased open days (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013). In contrast, other studies mention that inbreeding had no significant effect on specific traits such as birth weight and calving ease (McParland et al., Reference McParland, Kearney and Berry2009). We consider that the negative impact generated by inbreeding on most reproductive traits is evident. However, these effects would be presented in greater or lesser proportion to the levels of inbreeding present in each individual studied and directly proportional to the number of genomic regions harboring recessive mutations (Fritz et al., Reference Fritz, Capitan, Djari, Rodriguez, Barbat, Baur, Grohs, Weiss, Boussaha, Esquerré, Klopp, Rocha and Boichard2013) and lethal recessive mutations (VanRaden and Miller, Reference VanRaden and Miller2006). These effects have been increasing simultaneously with the evolution of higher yields in dairy cattle (Cole et al., Reference Cole, Null and VanRaden2016). In this sense, from the genotyping of genomic regions affected by inbreeding, further studies are suggested to identify a more significant proportion of regions to propose strategies to reduce the effects of inbreeding and maintain a balance between production levels, reproduction and inbreeding coefficients.
Influence of inbreeding depression in health and welfare
Health indices are fundamental factors in dairy farming as they directly influence animal welfare. In recent years, genomic evaluations of dairy cattle have incorporated new traits related to animal welfare that had not previously been considered. These traits play a major role in the overall health of the individual. In that sense, several studies give importance to the effect of inbreeding depression on health and animal welfare traits associated with dairy cattle longevity (Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019).
An increment of the inbreeding coefficient is a serious factor that increases the risk of activation of harmful recessive genes potentially influencing the cattle's immune system (Macedo et al., Reference Macedo, Bittar, Bass, Ronda, Bittar, Panetto, Araujo, Santos and Martins-Filho2014). Thus, increased inbreeding coefficients would be directly associated with the expression of recessive genes that impair the overall immune system, even affecting mitochondrial DNA (Macedo et al., Reference Macedo, Bittar, Bass, Ronda, Bittar, Panetto, Araujo, Santos and Martins-Filho2014; Baes et al., Reference Baes, Makanjuola, Miglior, Marras, Howard, Fleming and Maltecca2019), udder health (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019), linear conformation traits, dairy temperament, survival, general health status, individual vigor and strength (Cassell et al., Reference Cassell, Adamec and Pearson2003). Doekes et al. (Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019) also observed that inbreeding depression due to recent, compared to old inbreeding, significantly affected udder health. Health and animal welfare traits are strongly linked to the longevity of dairy cows, thereby increasing the time they remain in dairy operations, which impacts profitability. Therefore, these desirable health trait genotypes should be included in all dairy cattle evaluation, selection, and breeding systems to transmit them to future generations. We also emphasize that those dairy systems that maintain longevity traits in present populations will be highly profitable in the medium and long term.
Consequences of inbreeding depression on linear conformation
There is still some controversy regarding the impact of inbreeding on the conformation of dairy cows. The highly specialized conformation in high-yield dairy cattle has demanded the use of strongly related genetic lines initially derived from only about ten sires (Battagin et al., Reference Battagin, Sartori, Biffani, Penasa and Cassandro2013). Several authors claim that in dairy breeds such as Holstein, the high levels of inbreeding brought about by the convergence of genetic lines negatively affect several traits including linear conformation (morphological lines) (Croquet et al., Reference Croquet, Mayeres, Gillo, Vanderick and Gengler2006; Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010; Battagin et al., Reference Battagin, Sartori, Biffani, Penasa and Cassandro2013). These traits would be strongly associated with longevity, production, and reproductive capacity (Smith et al., Reference Smith, Cassell and Pearson1998; Sewalem et al., Reference Sewalem, Kistemaker, Miglior and van Doormaal2006). The reason for this is because adaptability traits, including conformation, longevity, and even disease resistance traits, show low heritability and decrease as levels of milk production per cow increase over time (Mirkena et al., Reference Mirkena, Duguma, Haile, Tibbo, Okeyo, Wurzinger and Sölkner2010). However, the importance of these characteristics derived from high inbreeding could be genotypically and phenotypically more related to longevity, production, and reproductive ability (Sewalem et al., Reference Sewalem, Kistemaker, Miglior and van Doormaal2006) than conformation. Another study showed that the degree of inbreeding had little effect on conformation traits but more adverse effects on production traits (Smith et al., Reference Smith, Cassell and Pearson1998). However, several studies based on a genealogical analysis in Holstein sires and dams regarding linear conformation traits showed that the degree of inbreeding significantly influences height, chest width, body depth, udder size and height, median suspensory ligament, udder depth, and teat placement (Rokouei et al., Reference Rokouei, Vaez Torshizi, Moradi Shahrbabak, Sargolzaei and Sørensen2010).
Repercussions of inbreeding depression on adaptability
The adaptability of dairy cattle to different environments has been the result of century-old natural evolution. Therefore the real adaptability of cattle to different current production systems and new environments is debatable. For example, a study conducted in nine native Russian cattle breeds identifying candidate known genes and other new genes related to domestication and production traits such as milk production (e.g. DGAT1, ABCG2), growth (e.g. XKR4), reproduction (e.g. CSF2) and environmental adaptation (e.g. AQP5, RAD50, RETREG1) showed highly variable results (Yurchenko et al., Reference Yurchenko, Daetwyler, Yudin, Schnabel, vander Jagt, Soloshenko, Lhasaranov, Popov, Taylor and Larkin2018). This way, during the last centuries, human beings have constantly been working out the specialization of dairy cattle to obtain a high-performance phenotype in the process of continuous matings of genetically related lineages with the accompanying increase in inbreeding rates (Refoyo-Martínez et al., Reference Refoyo-Martínez, da Fonseca, Halldórsdóttir, Árnason, Mailund and Racimo2019). Therefore, evaluating the individual inbreeding coefficient is crucial for the progress of animal adaptation, evolution, and conservation biology (Kardos et al., Reference Kardos, Luikart and Allendorf2015). Consequently, the degree of endogamy generated and the specialization of dairy cattle reached so far could have a considerable influence on the capacity of adaptability of dairy cattle to new environments and different production systems, for which additional research is necessary.
The breeding success and the improved adaptability of dairy cattle to different environments is controversial as this trait has low heritability (Strandén et al., Reference Strandén, Kantanen, Russo, Orozco-terWengel and Bruford2019). This natural evolutionary trait takes several centuries to become fixed in cattle (Åby and Meuwissen, Reference Åby and Meuwissen2014). Yet, the specialization of dairy breeds has led to an increase in inbreeding rates (Refoyo-Martínez et al., Reference Refoyo-Martínez, da Fonseca, Halldórsdóttir, Árnason, Mailund and Racimo2019), with variable heritability estimates observed regarding genes of domestication, production, reproduction, growth, and environmental adaptability (Yurchenko et al., Reference Yurchenko, Daetwyler, Yudin, Schnabel, vander Jagt, Soloshenko, Lhasaranov, Popov, Taylor and Larkin2018). The adequate and permanent monitoring of both individual and population inbreeding in dairy cattle is necessary to mitigate detrimental genetic effects related to cattle adaptability to different environments and production systems in the near future (Kardos et al., Reference Kardos, Luikart and Allendorf2015).
Accordingly, the need for the adaptation of dairy cattle to imminently emerging environments is a current concern. A recent study compared selection strategies in dairy cattle correlating productive traits (moderately heritable) with adaptive traits (low heritability) using simulations (Strandén et al., Reference Strandén, Kantanen, Russo, Orozco-terWengel and Bruford2019). This study concluded that genomic introgression (gene movements) produced more positive genetic change for both production and adaptive traits depending on the weight given to the adaptive or production traits during selection. Furthermore, this genomic introgression system seems to generate a lower risk of inbreeding. In general, results from simulations suggest that genomic selection can effectively introgress a low heritable trait into a target high-production population when the traits, i.e. the introgressed trait and production, are polygenic and genetically non-correlated (Åby and Meuwissen, Reference Åby and Meuwissen2014; Gaspa et al., Reference Gaspa, Veerkamp, Calus and Windig2015). Therefore, in the face of constant environmental changes that characterize current times, it is essential to highlight that one solution would point to introducing local adaptive genes in the most important dairy breeds (Nardone et al., Reference Nardone, Ronchi, Lacetera and Bernabucci2006; Hoffmann, Reference Hoffmann2010) (Berman, Reference Berman2011; Hoffmann, Reference Hoffmann2013). This genetic input could be an efficient strategy to introduce adaptive traits into commercial breeds (Strandén et al., Reference Strandén, Kantanen, Russo, Orozco-terWengel and Bruford2019; Hoffmann et al., Reference Hoffmann, Miller and Weeks2021).
It is also important to remark that dairy cattle populations must be prepared for future adaptive scenarios arising from the ongoing climate change. Therefore, the adaptability of animal populations to future environments will be vital for the proper performance of dairy breeds, suggesting that both adaptive and production traits can be simultaneously improved through genomic introgression (Strandén et al., Reference Strandén, Kantanen, Russo, Orozco-terWengel and Bruford2019). Therefore, introducing adaptive genes from donor dairy breed populations into less adapted recipient populations should be a central strategy. This introduction of genes would be a compelling way to increase genetic variability and avoid an increase in inbreeding depression and associated adverse effects on different genomic and phenotypic traits.
Strategic priorities to control inbreeding depression
Considering the increase in the annual loss of gene variability in studies involving Holstein sires (Doublet et al., Reference Doublet, Croiseau, Fritz, Michenet, Hozé, Danchin-Burge, Laloë and Restoux2019), recent work proposes that inbreeding should be controlled through evaluations based on genotyping because this control will be much more rigorous in genomic-based breeding programs than in those using pedigree for this purpose (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017; Mäntysaari et al., Reference Mäntysaari, Koivula and Strandén2020). Genomic selection has become an established approach and the standard in numerous dairy cattle production systems (Wiggans et al., Reference Wiggans, Cole, Hubbard and Sonstegard2017). Likewise, genomics has now paved the way for an extraordinary increase in genetic progress for most dairy cattle populations by drastically reducing the generation interval (Maltecca et al., Reference Maltecca, Tiezzi, Cole and Baes2020). Under genomic selection systems, an increase in the annual cumulative inbreeding rate has been observed due to the shortening of the generation interval through the use of genetic material from genetically related sires (Maltecca et al., Reference Maltecca, Tiezzi, Cole and Baes2020). This increase has favored the homozygosity effect to rapidly build up in dairy cattle populations (Howard et al., Reference Howard, Pryce, Baes and Maltecca2017). Therefore, a better understanding of how homozygosity and recessive load are related will ensure continuous genetic improvement, reduce the accumulation of harmful recessive genes and maintain genetic variability at the population level in dairy cattle (Maltecca et al., Reference Maltecca, Tiezzi, Cole and Baes2020). Regrettably, the accumulation of inbreeding depression is the unintended result of how selection has been performed in breeding programs over the past decades (Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019). Thus, the inbreeding rate per generation could decrease, since genomic selection can be used in a larger pool of candidates as we are not restricted by the limitations of traditional progeny testing systems, which allows only the testing of fewer candidates (Daetwyler et al., Reference Daetwyler, Villanueva, Bijma and Woolliams2007; Yurchenko et al., Reference Yurchenko, Daetwyler, Yudin, Schnabel, vander Jagt, Soloshenko, Lhasaranov, Popov, Taylor and Larkin2018). The degree of inbreeding is an imperfect measure of an individual's underlying recessive load because the accumulation of homozygosity for beneficial variants, compared to neutral or deleterious loci, cannot be distinguished (Gulisija and Crow, Reference Gulisija and Crow2007). However, information derived from the current genomic analysis methods has identified 18 lethal recessive loci (Cole et al., Reference Cole, Null and VanRaden2016). Therefore, the possibility to identify such loci with high accuracy may prevent adverse phenotypic effects in future matings, avoiding high inbreeding rates by identifying partial dominance and partial recessives (Jiang et al., Reference Jiang, Ma, Prakapenka, VanRaden, Cole and Da2019).
Estimates of future inbreeding evaluated through pedigree are more subjective and show a greater degree of underestimation (Sell-Kubiak et al., Reference Sell-Kubiak, Czarniecki and Strabel2018). However, the fact that genomic evaluation of inbreeding is more accurate than inbreeding levels obtained from pedigrees suggests that genomic inbreeding could better predict the actual recessive load to be taken into account in different dairy cattle breeding programs (Forutan et al., Reference Forutan, Ansari Mahyari, Baes, Melzer, Schenkel and Sargolzaei2018). Therefore, it is essential to highlight the need for the association of genomic and pedigree data to improve the estimation in the prediction of the traits, allowing us to perform matings with open lines of sires and consequently decreasing the degree of inbreeding (Gutiérrez-Reinoso et al., Reference Gutiérrez-Reinoso, Aponte, Cabezas, Rodriguez-Alvarez and Garcia-Herreros2020; VanRaden, Reference VanRaden2020).
Several state-of-the-art methods, such as those based on high-density single nucleotide polymorphisms (SNPs), are currently emerging as tools for identifying recessive lethal load, and therefore, for the selection of individuals carrying recessive genes (Cole et al., Reference Cole, Null and VanRaden2016; Maltecca et al., Reference Maltecca, Tiezzi, Cole and Baes2020) (Fig. 2). This fact could be considered a possible complementary tool to genome editing when variants are recessive (Cole, Reference Cole2015). However, a better formulation of different models, including dominance, has recently been achieved. This approach makes dominance estimates free of harmful effects arising from inbreeding (Vitezica et al., Reference Vitezica, Legarra, Toro and Varona2017).

Fig. 2. Main methods for the determination of inbreeding. The inbreeding coefficient (F) has been estimated from the pedigree of animals since around 100 years ago. A = animal = offspring; a = additive genetic relationship. FA →[0–1], is the probability for each locus of the offspring evaluated to be homozygous because their parents received the same alleles from a common ancestor (Oldenbroek and Waaij, Reference Oldenbroek and Waaij2014) 1; (Young and Seykora, Reference Young and Seykora1996)2. Genomic methods are based on SNPs (Single nucleotide polymorphisms) and provide higher accuracy to F estimation. Besides, the pedigree of animals does not need to be known. F PH = percentage of homozygosity of all SNP. N AA, N AB, N BB = number of SNP classified as AA, AB and BB, respectively. F PH does not distinguish between IBD (Identical by Descent, which what we aim F to be based on, represented by alleles descended from a common ancestor in a base population and IBS (identical by state, identical alleles regardless of whether they are inherited by a recent ancestor or not) (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013)3 G = matrix that contains allele frequencies, in rows → 0 – 2p (homozygotes); 1 – 2p (heterozygotes); 2 – 2p (opposite homozygotes). Columns correspond to each marker. p = allele frequency (VanRaden et al., Reference VanRaden, Olson, Wiggan, Cole and Tooker2011)4. ROH = Runs of homozygosity. F ROH can be estimated genomically. ROHs are a specific number of consecutive homozygous SNP. Inbreeding is characterized by high homozygosity and is highly clustered in the genome sequence space. Therefore, ROHs are long runs of homozygous SNP that become inherited together (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013)3. FROH is more accurate in the sense that it better detects IBD. FROH is higher when ROHs are long, and this will be associated with a short distance to common ancestors in the pedigree line (chromatin will not have had time to fragment during meiosis through more generations as would happen when related animals are further back in the pedigree), (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013)3; ![]() $\sum _k{\rm length}$ = number of ROH discoveries per animal; L = Total genome length (in kilobases, Kb). References: (1) (Oldenbroek and Waaij, Reference Oldenbroek and Waaij2014) (2) (Young and Seykora, Reference Young and Seykora1996) (3) (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013) (4) (VanRaden et al., Reference VanRaden, Olson, Wiggan, Cole and Tooker2011).
$\sum _k{\rm length}$ = number of ROH discoveries per animal; L = Total genome length (in kilobases, Kb). References: (1) (Oldenbroek and Waaij, Reference Oldenbroek and Waaij2014) (2) (Young and Seykora, Reference Young and Seykora1996) (3) (Bjelland et al., Reference Bjelland, Weigel, Vukasinovic and Nkrumah2013) (4) (VanRaden et al., Reference VanRaden, Olson, Wiggan, Cole and Tooker2011).
On dairy farms in different countries, including the USA, inbreeding control programs have been implemented at the population level using projections of expected future inbreeding or future genomic inbreeding (Sun et al., Reference Sun, VanRaden, Cole and O'Connell2014). However, one of the most effective methods for managing long-term genetic variability and inbreeding is optimum contribution selection (OCS). This has been available since the 1990s, although the practical use of this tool has been minimal (Maltecca et al., Reference Maltecca, Tiezzi, Cole and Baes2020). Therefore, it is essential to consider which selection allocates the contributions of each potential parent to minimize the overall weighted co-ancestry between prospective parents weighted by their contributions (Meuwissen, Reference Meuwissen1997).
One of the most important advances obtained to date has been the implementation of several computer methods such as the genomic best linear unbiased predictor (BLUP) of a single step (ssGBLUP) to perform studies of genome-wide association (GWAS), genomic prediction and estimation of several traits, taking into account the inbreeding index (Strandén et al., Reference Strandén, Matilainen, Aamand and Mäntysaari2017; Kang et al., Reference Kang, Ning, Zhou, Zhang, Yan and Liu2018). It has also been shown that the use of genomic relationships for inbreeding control is more efficient when assessing very large families of full siblings as opposed to single individuals (Clark et al., Reference Clark, Kinghorn, Hickey and Van Der Werf2013). Thus, runs of homozygosity (ROHs) have been proposed as a measure to track autozygosity and recent inbreeding that would be related to the actual recessive load of individuals (Clark et al., Reference Clark, Kinghorn, Hickey and Van Der Werf2013; Doekes et al., Reference Doekes, Veerkamp, Bijma, De Jong, Hiemstra and Windig2019) (Fig. 2).
Conclusions and future perspectives
Although genome editing technologies facilitate efficient cattle breeding without introducing transgenes, it can be difficult for the public to understand their possible benefits. However, we are currently witnessing that the use of strategies to eliminate deleterious alleles by genome editing is becoming real in cattle breeding, and it promises to be very advantageous. Thus, genome editing could prove helpful in eliminating deleterious recessive alleles identified by bioinformatics screening methods, which use sequence conservation and prior biological information about protein function. Genomics as a tool to control the harmful effects of inbreeding depression will continue to be the methodology of reference in the coming years. It will be increasingly used as an efficient set of methods to control and restrict the accumulation of genome homozygosity in dairy cattle. Breeding programs aim to maintain genetic diversity and limit the buildup of inbreeding, thus maximizing the response to selection. This goal can be achieved by maximizing the effective population size and minimizing the rate of inbreeding through control methods. With the adoption of genomics, the dairy genetics industry is slowly transforming and moving toward scenarios similar to those of other species in which control of population size is critical for inbreeding control in contrast to the information obtained from pedigree data. The combined use of data from pedigree-based relationships, genomics, OCS and ROH, among other methods, will help to improve accuracy at identifying the different lethal recessive loci of full and partial dominance, as well as other loci affecting other full and partial recessive traits. Also, inbreeding more closely related to overall recessive load could be determined either through the use of ROH or age-related partial inbreeding coefficients. Although managing lethal mutations has become more effective in recent years, considerable economic losses are associated with partial recessive loci of small effect. The significant amount of information gathered in recent years, based on several million individuals genotyped, offers an excellent opportunity to investigate partial recessive load and functional inbreeding depression, thus discriminating homozygosity and its potentially detrimental effects. The identification of truly deleterious partial recessives remains a long-term challenge in dairy cattle. The preservation of genetic diversity in future generations of dairy cattle is necessary to maintain current and future production in different environments. The application of intense genetic improvement and genetic introgression will guarantee increases in the frequency of favorable additive alleles and gene migration between donor and recipient populations. All these pieces of evidence suggest that genomic evaluation can be a standard tool in programs of reproduction, selection, and genetic improvement of dairy cattle.
Acknowledgements
We would like to thank the people and laboratories involved in the research related to the present review topic. Authors would like to acknowledge the support of the Universidad San Francisco de Quito (USFQ), Ecuador; Universidad Técnica de Cotopaxi (UTC), Ecuador; Agencia Nacional de Investigación y Desarrollo (ANID); Programa de Becas/Doctorado Nacional/2020–21201280, Chile and the National Institute for Agricultural and Veterinary Research (INIAV), Portugal. The contents of this review are solely the responsibility of the authors and do not necessarily represent the official views of the above-mentioned institutions.







