The replacement of fishmeal by plant protein sources is of great interest in aquafeed industry( Reference Fontainhas-Fernandes, Gomes and Reis-Henriques 1 , Reference Mbahinzireki, Dabrowski and Lee 2 ). However, approximately 70 % of the total P in plant feedstuffs is bound as phytate, which is poorly available for monogastric or agastric aquatic animals because of the absence of phytase in their gut for efficient phytate hydrolysis( Reference Jackson, Li and Robinson 3 , Reference Reddy, Sathe and Salunkhe 4 ). The presence of phytate in the aquafeed may also negatively affect growth performance, nutrient and energy utilisation and mineral uptake of fish( Reference Kumar, Sinha and Makkar 5 ). Furthermore, the undigested phytate-P ends up being excreted into the water, which may cause P pollution of the aquatic environment( Reference Cao, Wang and Yang 6 ).
Bound phytate-P can be effectively converted to available-P by phytase( Reference Cao, Wang and Yang 6 ). During the last decade, microbial phytase has been used by aquafeed industries to neutralise the negative effects of phytate( Reference Kumar, Sinha and Makkar 5 ). Dietary microbial phytase supplementation has been reported to improve the P bioavailibility( Reference Schafer, Koppe and Meyerburgdorff 7 – Reference Cao, Yang and Wang 13 ), mineral utilisation( Reference Jackson, Li and Robinson 3 , Reference Liebert and Portz 12 , Reference Van Weerd, Khalaf and Aartsen 14 – Reference Sugiura, Gabaudan and Dong 17 ), protein digestibility( Reference Schafer, Koppe and Meyerburgdorff 7 , Reference Furuya, Goncalves and Furuya 11 , Reference Oliva-Teles, Pereira and Gouveia 18 ) and growth performance( Reference Jackson, Li and Robinson 3 , Reference Vielma, Lall and Koskela 8 , Reference Liebert and Portz 12 , Reference Papatryphon and Soares 15 , Reference Debnath, Sahu and Pal 16 , Reference Vielma, Makinen and Ekholm 19 – Reference Baruah, Sahu and Pal 21 ) in different fish species. In addition, phytase can decrease P excretion into the aquatic environment( Reference Omogbenigun, Nyachoti and Slominski 22 ). In Nile tilapia, supplementation of microbial phytase at 1000 U/kg feed resulted in growth and mineral utilisation similar to that of a plant-based diet supplemented with inorganic P( Reference Liebert and Portz 12 , Reference Portz and Liebert 23 , Reference Liebert and Portz 24 ).
Studies about microbial phytase supplementation in aquafeed have mainly focused on the effect on nutrient utilisation and fish growth performance, the dose–response, efficacy of phytases from different sources and the most efficient ways of supplementation( Reference Kumar, Sinha and Makkar 5 , Reference Cao, Wang and Yang 6 , Reference Sajjadi and Carter 9 , Reference Furuya, Goncalves and Furuya 11 , Reference Liebert and Portz 12 , Reference Portz and Liebert 23 , Reference Nelson, Shieh and Wodzinski 25 ). However, as a feed-additive enzyme, microbial phytase may modulate the gut microbiota through the hydrolysis of phytate, and thus influence the gut health. Smulikowska et al. ( Reference Smulikowska, Czerwinski and Mieczkowska 26 ) reported higher levels of acetic and butyric acids in caecal contents of broiler chickens fed phytase, suggesting a modulation of the microbial activity due to more P and other nutrients released from phytates. In pigs, supplemental phytase was observed to increase the Clostridium group in the ileum without changing total bacterial numbers( Reference Metzler-Zebeli, Vahjen and Baumgartel 27 ). However, there has been no study about the effect of microbial phytase on gut health of fish. Therefore, the main purpose of our study was to evaluate the impact of microbial phytase on gut health of fish, focusing on the effect on intestinal histology, adhesive microbiota and the expressions of stress-related gene markers and inflammation-related cytokine genes. Hybrid tilapia (Oreochromis niloticus ♀×Oreochromis aureus ♂) was selected as the experimental target because of its strong environmental adaptability, ease of breeding and high socio-economic importance in both developing and developed countries( Reference Verdegem, Hilbrands and Boon 28 ).
Methods
Fish and diet
The microbial phytase (derived from strain Aspergillus niger 963, expressed by Pichia pastoris, 5×105 U/ml culture medium) was obtained from Challenge Group (Beijing, China). Juvenile hybrid tilapia (O. niloticus ♀×O. aureus ♂) were raised in the tilapia breeding base of Jiaxing (Zhejiang, China). The basal diet (diet 1) was formulated according to the recommendations for tilapia( 29 ), and the treatment group (diet 2) contained 1000 U/kg microbial phytase (supplementation of microbial phytase from 500 to 1500 U/kg, especially 1000 U/kg of feed, led to improved growth, mineral utilisation and protein digestibility in tilapia( Reference Kumar, Sinha and Makkar 5 , Reference Furuya, Goncalves and Furuya 11 , Reference Liebert and Portz 12 )). The pelleted diets (Table 1) were produced by a small pellet mill to minimise the impact of mixing and granulation (high temperature process) on phytase activity. The enzymatic activity of phytase supplemented in feed was checked before and after the feeding period according to the method of Engelen et al.( Reference Engelen, van der Heeft and Randsdorp 30 ).
Table 1 Diet formulations and their chemical compositions
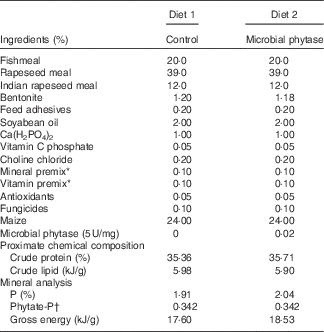
* National Research Council( 29 ).
† The phytate-P was estimated by the generally known phytate-P content in maize (0·15 %) and rapeseed meal (0·6 %).
Fish husbandry
In total, 120 healthy tilapia were acclimated for 1 month before the experiment. After the acclimation period, tilapia (19·3 (se 0·02) g) were weighed and randomly distributed to two experimental groups with four replicates, and then reared for 12 weeks in cages (3×3×2 m). Fish were hand-fed to apparent satiation twice a day for 12 weeks. The feeding rate was approximately 3 % at the beginning of the trial, and thereafter was increased weekly by weighing all fish in one cage per treatment. At each meal, fish in each cage were fed multiple times until apparent satiation within 30 min, with no or negligible remains. Water quality parameters were checked each day. Temperature was in the range of 27·1–36·5°C and pH was 7·6 (se 0·1); ammonia and nitrite-nitrogen were maintained under 0·2 mg N/l and 0·05 mg N/l, respectively. At the end of the experiment, the fish were fasted for 24 h before sampling and harvest. All experimental and animal care procedures were approved by Feed Research Institute of Chinese Academy of Agricultural Sciences Animal Care Committee, under the auspices of the China Council for Animal Care (assurance no. 2009-HSX01). MS-222 was used as the anaesthetic. The field study was conducted in a private aquaculture pond with the owner’s permission.
Production and phosphorus utilisation
Total number and body weight of fish in each cage were measured at the beginning and at the end of the 12-week feeding period. Weight gain (WG), food conversion ratio (FCR) and survival rate were assessed using the following formulae:
Fish were killed using 25 mg/l of tricaine methanesulphonate (MS-222) before sampling. Bone and scale samples were obtained from killed fish. Bones from ten fish were collected, boiled in de-ionized water for 2 min and cleaned thoroughly to remove soft tissues. Scales were collected from both lateral surfaces of ten fish and mucus was wiped off. Next, three samples were randomly chosen and pooled as one sample. All samples were maintained at –70°C before analysis( Reference Hughes and Soares 31 ). Total P in feed, bone and scale samples were determined using the vanadium–molybdate method according to the modified Association of Official Analytical Chemists (AOAC) method 986.11. The absorbance was measured at 640 nm using a UV–Vis spectrophotometer (Specord S100; Carl Zeiss)( Reference Liebert and Portz 24 ). The phytate-P in feed was estimated by the generally known phytate-P content in rapeseed meal (0·6 %) and maize (0·15 %).
Gut histology
Intestinal samples from three fish per cage (n 8) were obtained and fixed using 2·5 % glutaraldehyde for scanning electron microscopy (SEM) studies, according to the method described by Merrifield et al.( Reference Merrifield, Dimitroglou and Bradley 32 ). Micrographs (magnification 100×) were used to observe the intestinal mucosa folds, and SEM micrographs (magnification 20 000×) were analysed to measure microvilli length( Reference Fagundes-Neto, De Martini-Costa and Pedroso 33 ). SEM micrographs were analysed using ImageJ 1.36 (National Institutes of Health).
Gut-adhesive bacteria
Three cages of each group were randomly selected and three fish per cage were dissected under MS-222 (25 mg/l) anaesthesia. Intestines were aseptically removed, opened and gently agitated three times for 1 min in PBS to remove the contents and then pooled to investigate gut-adhesive bacteria( Reference Ringø 34 ). Genomic DNA was obtained using the extraction method as described by He et al.( Reference He, Zhou and Liu 35 ) with some modifications. In brief, 200 mg tissue samples were homogenised in 500 ml lysozyme lysis buffer (0·3 m-sucrose, 0·025 m-EDTA, 0·025 m-Tris-HCl, pH 8·0) and incubated at 37°C for 1 h. The samples were gently inverted every 15 min. Next, 1 ml CTAB lysis buffer (0·1 m-Tris-HCl, 0·1 m-Na-EDTA, 1·5 m-NaCl, 1 % CTAB, 2 % SDS, pH 8·0) was added, and the samples were further incubated at 65°C for 4 h, with gentle inversion every 15 min. After incubation, the samples were centrifuged at 13 000 g for 10 min. The supernatant was transferred to sterilised tubes and an equal volume of trichloromethane was added, followed by gentle inversion and centrifugation at 13 000 g for 10 min. The supernatant was transferred into a new tube and gently mixed with equal volume of isopropanol and precipitated at −20°C for 30 min, followed by centrifugation at 13 000 g for 10 min. The pellet was collected, washed with 75 % ethanol and re-suspended in 50 μl double-distilled water. Genomic DNA was thereafter purified using a TIANquick Midi purification kit (TIANGEN).
The V3 region of the 16S ribosomal RNA (rRNA) gene was amplified with primers 338-GC-f and 519-r (Table 2). PCR-DGGE (denaturing gradient gel electrophoresis) was performed according to Liu et al.( Reference Liu, Zhou and Yao 36 ) and Zhou et al.( Reference Zhou, He and Liu 37 ) using a DCode universal Mutation System (Bio-Rad). Purified PCR products were loaded onto denaturing gradient gels ranging from 40 to 60 % polyacrylamide, and then electrophoresis was carried out at a constant voltage of 60 V for 16 h in 1×Tris-acetate-EDTA buffer at 60°C. The gels were stained in ethidium bromide solution (0·5 mg/ml in Tris-acetate-EDTA buffer), destained in distilled water for 20 min and viewed under UV transillumination. The dominant bands were excised, re-amplified, purified (TIANquick Midi purification kit; TIANGEN) and sequenced. Computer-assisted comparison of DGGE patterns was performed with Quantity One version 4.62 (Bio-Rad).
Table 2 Primers used in this study

rRNA, ribosomal RNA.
Expression of hsp70 and immune-related cytokine genes
The intestinal expression of hsp70 and immune-related cytokine genes was assessed. Intestinal tissues were sampled from three fish per cage at the end of the 12-week feeding period. All the tissues from the same group were pooled and frozen in liquid N2 and stored at −70°C. Total RNA was extracted using a TRIzon Reagent RNA kit (Promega). The integrity of total RNA was verified by visualisation on 1·2 % agarose gel. RNA was dissolved in 50 ml RNase-free water and stored at −70°C until use. Complementary DNA (cDNA) was synthesised for quantitative RT PCR (RT-qPCR) using the ReverTra Ace-a-RT-PCR kit (Toyobo).The qPCR primers were referenced from Liu et al.
(
Reference Liu, Ren and He
38
) (Table 2). A sample of 2 μl of cDNA was amplified by forty cycles in a 20-μl reaction system with the SYBR Green Premix Ex Taq™ II (TaKaRa) in an iQ5 multicolour real-time PCR detection system (Bio-Rad). The PCR conditions were as follows: 95°C for 3 min, forty cycles (95°C 20 s, 58°C 20 s and 72°C 20 s), and an additional final extension at 72°C for 10 min. Data analysis was conducted using the
![]() $2^{{{\minus}\Delta \Delta C_{T} }} $
method. β-Actin and 18S rRNA were used as the reference genes. All real-time PCR were performed at least three times and data were analysed using Bio-Rad iQ5 software.
$2^{{{\minus}\Delta \Delta C_{T} }} $
method. β-Actin and 18S rRNA were used as the reference genes. All real-time PCR were performed at least three times and data were analysed using Bio-Rad iQ5 software.
Statistical analysis
Results are expressed as mean values with their standard errors. Statistical analyses were performed using GraphPad Prism 5. Differences between means were subjected to Student’s t test and were considered significant when probability (P) values <0·05 were obtained. Cluster analysis was based on the unweighted pair group method using the arithmetic mean algorithm (UPGMA). Similarity between bacterial communities was evaluated with a pairwise similarity coefficient (Cs, the measure of the similarity of two samples by UPGMA): <0·60 were regarded as different, those with 0·60≤Cs<0·80 were considered to be marginally different and those with Cs≥0·80 were considered to be similar( Reference Wang, Zhou and He 39 ).
Results
Production and phosphorus utilisation
WG and feed utilisation are displayed in Table 3. There were no significant differences between the two experimental groups (P>0·05). Feed used for the two groups had equal amounts of total P and phytate-P (Table 1). After the 12-week feeding period, the scale P content of tilapia was significantly higher than control (P<0·05) in the microbial phytase group, whereas bone P content was not influenced by phytase supplementation (Table 3).
Table 3 Growth-related parameters, survival rate and the percentage of phosphorus in bones and scales of tilapia fed the two experimental diets at the end of the feeding period (Mean values with their standard errors)
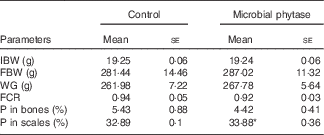
IBW, initial body weight; FBW, final body weight; WG, weight gain; FCR, feed conversion ratio.
* P<0·05 (Student’s t test).
Gut histology
SEM images showed that the arrangement of the intestinal mucosa folds in tilapia fed microbial phytase was more regular and tighter compared with the control (online Supplementary Fig. S1). The gut microvilli was significantly shorter (P<0·05) but the microvilli density was higher (P<0·05) in the microbial phytase group compared with the control (Table 4; online Supplementary Fig. S2).
Table 4 Intestinal microvilli length (by estimate) and density of tilapia fed the two experimental diets at the end of the feeding period (Mean values with their standard errors)
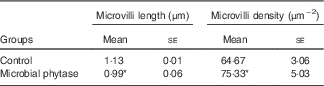
* P<0·05 (Student’s t test).
Gut-adhesive bacteria
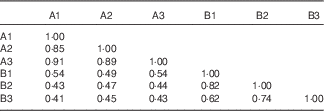
The 16S rRNA gene V3 PCR-DGGE fingerprints of the gut-adhesive bacterial communities for the two experimental groups are shown in Fig. 1. Different band patterns can be visually recognised. Closest relatives of representative bands in the PCR-DGGE fingerprints are illustrated in Table 5. Bands 1, 3, 5, 6, 7 and 15 showed high similarity (98–100 %) with uncultured bacteria, in which bands 1 and 3 represent the most predominant intestinal-adhesive bacteria in the control group, with the average relative abundance of 17·0 and 27·6 %, respectively. The relative abundances of bands 1 and 5 were drastically reduced or even disappeared in the phytase-supplemented group v. control, whereas the abundances of bands 3 and band 6 showed a moderate decrease and increase, respectively (P<0·01). Meanwhile, band 11, with 99 % similarity to Vibrio sp., and band 8, with 99 % similarity to Thermus sp., showed significant increases (P<0·05) in the phytase-supplemented group compared with control, whereas the intensity of band 16, with 99 % similarity to Exiguobacterium sp., significantly decreased. Bands 4, 9 and 12 were unique in the microbial phytase group, and were identified as Vibrio sp., Clostridium sp. and uncultured proteobacterium, respectively. At the phylum level, the collective abundance of bands belonging to Firmicutes and Thermus was significantly elevated by microbial phytase supplementation (P<0·01 for Thermus, P<0·05 for Firmicutes), whereas no significant difference was observed for the collective richness of both Proteobacteria and Actinobacteria. Both the cluster analysis (Fig. 2) and pairwise Cs matrix (Table 6) indicated significant influence of microbial phytase supplementation on gut-adhesive bacterial communities (Cs<0·60).

Fig. 1 16S ribosomal RNA gene V3 PCR-DGGE fingerprints of the gut-adhesive bacterial communities in tilapia of the control group and the microbial phytase group. A1, A2, A3: the control group; B1, B2, B3: the microbial phytase group. Number means different replicates.

Fig. 2 Cluster analysis of the gut-adhesive bacterial communities in tilapia of the control and microbial phytase groups based on 16S ribosomal RNA gene V3 DGGE fingerprints. A1, A2, A3: the control group; B1, B2, B3: the microbial phytase group.
Table 5 Representatives of the predominant gut-adhesive bacterial types in tilapia fed the two experimental diets as determined by the 16S ribosomal DNA V3 DGGE fingerprints (Mean values with their standard errors)

* P<0·05, ** P<0·01 (Student’s t test).
Table 6 Pairwise similarity coefficients matrix for the gut-adhesive bacterial communities of tilapia fed the two experimental diets at the end of the feeding period
Expression of hsp70 and immune-related cytokine genes
The relative expressions of gut immune-related cytokine genes (tnf-α, il-1β and tgf-β) and hsp70 were tested to evaluate the influence of microbial phytase supplementation on gut immunity and stress status of tilapia. The gut inflammatory responses were stimulated in the microbial phytase supplementation group, as indicated by the up-regulation of tnf-α and tgf-β (Fig. 3). The relative expression of hsp70 showed approximately 6-fold increase in the microbial phytase group compared with control (P<0·05), indicating an elevated gut stress status by the microbial phytase.

Fig. 3 Effects of dietary microbial phytase on intestinal cytokine gene expressions in tilapia at the end of the 12-week feeding period. Values are means (n 4), with standard errors represented by vertical bars. * P<0·05 (Student’s t test). ![]() , Control;
, Control; ![]() , microbial phytase.
, microbial phytase.
Discussion
Studies about microbial phytase supplementation in diets for fish have mainly focused on the effect on nutrient utilisation and fish growth performance( Reference Kumar, Sinha and Makkar 5 , Reference Cao, Wang and Yang 6 , Reference Sajjadi and Carter 9 , Reference Furuya, Goncalves and Furuya 11 , Reference Liebert and Portz 12 , Reference Portz and Liebert 23 ). No study has investigated the effect of microbial phytase on gut health of fish, which is important for fully evaluating the outcomes of phytase supplementation. In this study, the impact of microbial phytase on gut health of fish was evaluated, focusing on the effect on intestinal histology, adhesive microbiota as well as the stress and inflammation status in the gut.
Both dietary groups grew well with high body WG after 12 weeks of feeding and there were no deaths throughout the experiment in both groups. Microbial phytase supplementation at 1000 U/kg did not improve tilapia’s production at a statistically significant level, and only a trend of improvement was observed. However, positive impacts of dietary microbial phytase on tilapia growth have been reported before. Diets with 1000 U/kg phytase supplementation can significantly improve growth of Nile tilapia( Reference Portz and Liebert 23 ). Moreover, Cao et al.( Reference Cao, Yang and Wang 13 ) reported that pre-treatment of plant ingredients with phytase at 1000 U/kg had significantly positive effects on growth performance in Nile tilapia. The discrepancy of our result with previous ones might be attributed to the differences in the basal diet formulation, source of microbial phytase as well as rearing conditions compared with previous studies( Reference Furuya, Goncalves and Furuya 11 ). Our preliminary 4-week feeding experiment in tilapia, with diets supplemented with currently used microbial phytase at 0, 500, 1000, 2000 and 4000 U/kg, showed that only the dose of 1000 U/kg resulted in a trend in growth improvement and higher doses showed decreased growth compared with 1000 U/kg (data not shown here). Therefore, the dose effect should not be a factor responsible for the observed insignificant improvement in growth performance of tilapia.
Furuya et al.( Reference Furuya, Goncalves and Furuya 11 ) observed that phytase supplementation between 500 and 1500 phytase units/kg improved P availability for tilapia. Portz & Liebert( Reference Portz and Liebert 23 ) reported that microbial phytase supplementation may improve P content in the vertebra and scales of tilapia. In our study, the P content in bones was not affected by phytase, but there was a significantly higher level of P content in the scales of tilapia fed the microbial phytase diet. As we know, P level in scales but not in the vertebrae was more sensitive to dietary Ca( Reference Ye, Liu and Tian 40 ), Cd( Reference Berntssen, Waagbø and Toften 41 ) and also other elements( Reference Lall 42 , Reference Lall and Lewis-McCrea 43 ). Moreover, scales are the source of reserve P in fish. Available P is first utilised by the bone for proper mineralisation and then utilised for scale growth and storage. Scale P appears to be easily mobilised for utilisation in the body when needed and is closely related to dietary P status( Reference Hughes and Soares 31 ). Therefore, these results indicate the efficiency of the microbial phytase to improve P bioavailability in our study.
The effects of phytase supplementation on gut bacteria of fish are largely unknown, despite their critical role in host nutrition, health and production( Reference Fuller, Perdigón and Fuller 44 ). In this study, a significant impact of microbial phytase on the gut microbial community was noted based on 16S rRNA gene DGGE fingerprints. Evidences suggest that diet composition is a strong modulator of host gut microbial composition( Reference Kiarie, Romero and Nyachoti 45 ). Therefore, the influence of phytase on the gut microbiota might be due to the increased supply of dissolved P and other nutrients by the hydrolysis of phytate( Reference Cao, Wang and Yang 6 ). The relative abundance of Clostridium sp. and Thermus sp. was significantly stimulated by microbial phytase supplementation. Notably, Clostridium sp. may ferment carbohydrates into SCFA such as butyrate and propionate, which play an important role as fuel for intestinal epithelial cells( Reference Fuller, Perdigón and Fuller 44 , Reference Vinolo, Rodrigues and Nachbar 46 ) and anti-inflammatory factors( Reference Atarashi, Tanoue and Oshima 47 ). Similar to our result, Smulikowska et al. ( Reference Smulikowska, Czerwinski and Mieczkowska 26 ) reported that the villus length and crypt depth in the jejunum and ileum of broiler chicken were increased by dietary phytase supplementation, and they attributed these effects to the higher production of SCFA due to the modulation of gut microbiota and the increase in their metabolic activity.
In this study, the microvillus length and density were used as the morphology parameters to evaluate intestinal health. The gut mucosa microvilli densities were significantly enhanced by phytase supplementation. However, gut microvilli length was inhibited, indicating that microbial phytases have both beneficial and detrimental effects on gut morphology.
Heat shock protein 70 (hsp70) played essential roles in protein metabolism under normal and stress conditions, including de novo protein folding, membrane translocation, degradation of misfolded proteins and other regulatory processes( Reference Iwama, Thomas and Forsyth 48 , Reference Xing, Li and Wang 49 ). The expression of hsp70 gene was used as the biomarker of host stressful conditions in certain organs( Reference Morimoto 50 ). Inflammatory cytokines including il-1β, tnf-α (pro-inflammatory factors)( Reference Kudo, Fujikawa and Itonaga 51 ) and tgf-β (anti-inflammatory factor)( Reference Sano, Shimizu and Sato 52 ) are important mediators of inflammation released by activated phagocytes and are commonly used as reference genes in studies of inflammatory status. The expression of hsp70 in the kidney of tilapia was significantly reduced (P<0·05) in the phytase group (online Supplementary Fig. S4), which may reflect a general improved welfare of fish fed microbial phytase. However, the expressions of hsp70, tnf-α and tgf-β in the gut of tilapia were increased (Fig. 3), which revealed a higher gut stress and inflammatory status induced by microbial phytase. To our knowledge, this is the first report to show that phytase supplementation may lead to enhanced inflammation and stress in the gut of fish, which might act as an additional reason contributing to the failure of phytase to improve the overall growth performance of tilapia in this study, as the increased energy expenditure caused by gut inflammation( Reference Qiu, Croom and Ali 53 ) might counteract the positive effects of dietary microbial phytase on growth. In addition, the enhanced gut inflammation status might be responsible for the decreased microvilli length.
Taken together, this microbial phytase exerted mixed effects on tilapia in terms of gut health, that is, improved microvilli density v. reduced microvilli length and higher stress/inflammation status in the gut. The negative effects of phytase might be due to the following reasons: (1) the direct adverse impacts to host fish by this microbial phytase or/and (2) the indirect effects to host fish by the change in gut microbiota induced by the dietary phytase. As mentioned above, the enrichment of Clostridium sp. in the gut microbiota of fish fed phytase may implicate an elevated SCFA level in the gut, which should benefit the gut morphology( Reference Fuller, Perdigón and Fuller 44 , Reference Vinolo, Rodrigues and Nachbar 46 ) and inflammatory status( Reference Atarashi, Tanoue and Oshima 47 ). Therefore, the indirect effect mediated by change in microbiota was putatively beneficial, and the observed adverse impacts were probably attributed to the direct effect of microbial phytase on the host, which deserves further investigation by experiments in germ-free fish models.
Some exogenous enzymes may exert adverse effects on fish production. Kazerani et al. ( Reference Kazerani and Shahsavani 54 ) observed a dose-dependent reduction in the growth performance of common carp (Cyprinuscarpio) fed commercial exogenous multienzymes containing xylanase and β-glucanase. The authors hypothesised that these exogenous enzymes may result in the production of galactose and xylose from dietary NSP in the intestine of fish, to which most fish species are intolerant to( Reference Stone, Allan and Anderson 55 ). Similarly, Mahmoud et al.( Reference Mahmoud, Kilany and Dessouki 56 ) observed that commercially prepared exogenous multienzyme preparations Pan Zyme (containing xylanase, acidic proteinase, neutral proteinase and cellulase) and Phytase-plus broiler 500 (containing phytase enzyme; Bytara) both reduced the production of Nile tilapia. The Pan Zyme group showed mild degeneration of intestinal mucosa as well as focal detachment of the epithelial lining, whereas Phytase-plus broiler 500 led to degeneration of intestinal mucosa and mild enteritis characterised by aggregation of mononuclear cells (mainly lymphocytes). In the present study, the microbial phytase reduced the intestinal microvilli height and increased the intestinal inflammation and stress status of host fish, which might be due to its antigenicity.
In conclusion, dietary microbial phytase may exert mixed effects on hybrid tilapia, and could guide our future selection of phytases as aquafeed additives, that is, eliminating those that can stimulate intestinal inflammation.
Acknowledgements
The authors are indebted to Lei Zhu's help throughout the feeding experiment. The authors are grateful to Yang Deng for his excellent technical assistance.
This work was supported by the National Natural Science Foundation of China (31272672, 31572633), the Key Project of Chinese National Programs for Fundamental Research and Development (973 Program) (2015CB150605), and Beijing earmarked fund for Modern Agro-industry Technology Research System (SCGWZJ 20151104-4).
The authors’ contributions are as follows: J. H. performed all the data analysis and wrote a draft of the manuscript. Y. Y. and Y. C. managed the feeding trial and cytokine gene expression analysis. S. H. completed the DGGE and electron microscopy analysis. X. Z. and Z. Z. designed and guided the experimental performance and helped in the writing of the manuscript. C. R. and B. Y. improved the manuscript by critical comments and suggestions.
There are no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit http://dx.doi.org/doi:10.1017/S0007114516001240












