Introduction
Parasite remains – larvae, oocysts, cysts and, mainly, eggs – found in coprolites have mostly been attributed to helminths and protozoa from the gastrointestinal tract and, to a lesser extent, from the respiratory system (e.g. Eucoleus aerophilus) of the definitive host. In the last 15 years, we have performed several coproparasitological studies in archaeological sites from southern Patagonia in Argentina. These studies were mainly carried out in caves and focused on hosts: hunter gatherers and other mammals which took refuge in rockshelters, such as camelids, rodents, foxes and pumas (Fugassa et al., Reference Fugassa, Denegri, Sardella, Araújo, Guichón, Martinez, Civalero and Aschero2006; Amalfitano et al., Reference Amalfitano, Petrigh, Loos and Fugassa2017; Fugassa and Petrigh, Reference Fugassa and Petrigh2017). The characteristics of the ancient environments and epidemiological aspects guided our parasitological research in Patagonia. In this context, parasite assemblages were identified and compared discussing the local processes that configured them (Amalfitano et al., Reference Amalfitano, Petrigh, Loos and Fugassa2017; Fugassa and Petrigh, Reference Fugassa and Petrigh2017). In the last few years, ancient DNA (aDNA) analysis for coprolite identification was implemented. aDNA sequences were obtained from two coprolites of puma (Puma concolor) and one of red fox (Lycalopex culpaeus) from the Middle/Late Holocene (2700, 5100 and 2900 years old, respectively) (Petrigh and Fugassa, Reference Petrigh and Fugassa2017; Fugassa et al., Reference Fugassa, Petrigh, Fernández, Catalayud and Belleli2018). This approach complemented previous parasitological findings, supporting our assumptions. Although aDNA can be scarce due to damage accumulated over time (Paäbo et al., Reference Paäbo, Poinar, Serre, Jaenicke-Després, Hebler, Rohland and Hofreiter2004), the multiple copies of mitochondrial DNA and the favourable dry and cold conditions at the studied sites have made it possible to obtain nucleotide sequences of ancient remains (Briggs et al., Reference Briggs, Stenzel, Johnson, Green, Kelso, Prüfer and Pääbo2007).
The aim of the present work was to explore gastrointestinal parasites in coprolites from a distant and different context: Peñas de las Trampas 1.1 (PT1.1) paleontological and archaeological site in the southern Andean Puna of Argentina. Peñas de las Trampas rockshelter, just like the Patagonian sites previously studied, started its long occupational history in the Late Pleistocene period, with extinct megafauna as the earliest protagonists (Martínez, Reference Martínez and Elizabeth2014). In addition, some key aspects of PT1.1 and the Patagonian sites previously studied make them comparable, such as all of them being rockshelters and sharing the key mammalian species in the respective ecosystems. Indeed, both the Puna and Patagonia belong to the arid Andean–Patagonian sub-region of the Neotropics.
In this context, we conducted the first exploration of entero-parasites of Pleistocene carnivorous mammals of this Puna site, combining morphometric and molecular analyses to achieve coprolite and parasite identification. The largest carnivores in an ecosystem have been defined as particularly important species in its configuration and functioning (Miller et al., Reference Miller, Dugelby, Foreman, Del Río, Noss, Phillips and Willcox2001). In addition, carnivores are central due to their role in the dispersal of their own entero-parasites of direct and indirect cycles, as well as of the entero-parasites of their prey.
Materials and methods
Peñas de las Trampas 1.1 paleontological and archaeological site is in a rockshelter on top of a hill, with an opening of 23 m and a maximum depth of 10 m (Martínez, Reference Martínez and Elizabeth2014). It is located at 3582 m above sea level in Antofagasta de la Sierra, Catamarca Province, Argentina, in the southern Puna. Remains and traces of extinct megafauna including giant ground sloths (Megatheriinae and Mylodontinae gen. sp.) and Pleistocene horse (Hippidion sp.) have been found at this site. Although at present the area is extremely dry – 100 mm annual rainfall – the presence of mega-herbivores suggests that wetter environmental conditions prevailed until the Holocene, which were favourable to these populations. No association between extinct fauna and humans has been found so far, and archaeological occupations at the site and in the area are only documented, generally, since the very end of the Pleistocene (Martínez, Reference Martínez and Elizabeth2014; Mondini and Elkin, Reference Mondini, Elkin and Elizabeth2014).
One coprolite (catalogue No. 760, Laboratorio de Parasitología de Sitios Arqueológicos) from layer IV of PT1.1 was selected for this exploratory study. Stratigraphic layer IV was previously dated to 23 195–23 913 cal yr BP. A fraction of the analysed coprolite was submitted to radiocarbon dating by Accelerator Mass Spectrometry (AMS) at the Center for Applied Isotope Studies, University of Georgia (CAIS-UGA), USA. Calibration of dates was made with Calib 7.1 (2 sigma).
Parasitological examination was performed on the coprolite contents. First, it was rehydrated with sterile phosphate-buffered saline (PBS) 1x for 48 h at 4 °C, sieved in a 300 µm mesh and, subsequently, spontaneous sedimentation took place for 2 h. Twenty transient slides were made and remains retained in the mesh were preserved for future diet studies. The parasite eggs, identified under a light microscope (100× magnification), were manually isolated by the use of modified capillaries and stored in PCR tubes with 5 µL of PBS 1x at −20 °C. The molecular identification of the coprolite was performed with 50 mg by duplicates. Two coprolite fragments were separated into a horizontal laminar airflow workbench. DNA was isolated from duplicates and one negative control was added. Exclusive sterile reagents and disposable supplies and equipment were used for each process. Sample processing, DNA extractions and PCR amplifications were performed into horizontal laminar airflow workbenches located in exclusive areas for work in aDNA and separated from each other. Rooms, laminar airflow workbench and surfaces were decontaminated with 10% bleach and 70% ethanol and irradiated with UV light prior to use (Petrigh and Fugassa, Reference Petrigh and Fugassa2017; Fugassa et al., Reference Fugassa, Petrigh, Fernández, Catalayud and Belleli2018). A 170 bp of mitochondrial cytochrome b (cytb) gene was amplified by PCR using carnivore-specific primers (Farrell et al., Reference Farrell, Roman and Sunquist2000). Four PCR amplifications were performed to each coprolite fragment (eight replicates of the coprolite). DNA extraction, PCR setup and thermocycler program were those described in Petrigh and Fugassa (Reference Petrigh and Fugassa2017). Four negative controls were added in PCR experiments.
aDNA extraction and PCR amplification from Toxascaris sp. eggs were carried out following the procedures described in Petrigh et al. (Reference Petrigh, Scioscia, Denegri and Fugassa2015). Two independent DNA extractions of two egg pools (16 eggs of Toxascaris sp. each) were performed at different moments in time (11 months apart), and one negative control was added in each DNA extraction. Egg identification was performed by PCR amplification of a 151 bp cytochrome c oxidase I (cox1) gene fragment using Ascaris genus-specific primers: As-Co1F forward (Peng et al., Reference Peng, Yuan, Hu, Zhou and Gasser2005) and ACR1B reverse (Søe et al., Reference Søe, Nejsum, Fredensborg and Kapel2015). Several PCR amplifications were performed separately in time. The first experiment was performed in September 2017, the second in April 2018 and the third in March 2019. The first was performed with 14 replicates, and the second included 12 replicates. Four negative controls were added in the first and second PCR experiments. In the third PCR amplification, two DNA replicates (two PCR amplifications) and two negative controls were analysed. DNA sequencing was performed by the Sanger method at the Genomic Unit of Biotechnology Institute from the National Institute of Agricultural Technology (INTA Castelar), Buenos Aires, Argentina. Chromatograms were analysed with the BioEdit v7.0.5 program (Tom Hall, Ibis Therapeutics, 2005) and consensus sequences between replicates were built for each sample. Identification of sequences was conducted by the BLASTN algorithm of the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome).
Results
Coprolite No. 760 was dated to 16 573–17 002 cal yr BP with 95.4% probability (UGAMS 32587; 13 910 ± 30 yr BP). The radiocarbon dating of the coprolite thus rendered a younger age than that of layer IV, where it was contained. This disagreement could result from a vertical migration of the coprolite through the sedimentary layers, prompted, for example, by the circulation of the megafauna inside the rockshelter. This result highlights the importance of getting host taxon dates for coprolites themselves, which is rare in ancient parasite studies, so as to be able to determine their age accurately.
The coprolite had a maximum diameter of 25.4 mm (Fig. 1) and rounded ends, suggesting that it could correspond to a large canid or, most probably, to a large felid like a puma or a jaguar (Chame, Reference Chame2003). To test the hypothesis of its feline origin, aDNA analysis was conducted as described in Petrigh and Fugassa (Reference Petrigh and Fugassa2017) and above, applying specific protocols to minimize contamination risk with modern or exogenous DNA. A 170 bp fragment of cytb gene from carnivores was amplified by PCR in the two DNA replicates from the coprolite. Sequence chromatograms were typical of aDNA molecules with post-mortem damage, and consistent with the sample old age (e.g. baseline noise, a few double peaks with C/G and T/A by deamination) (Lindahl, Reference Lindahl1993). However, a reliable consensus sequence could be defined from both duplicates. These results highlight the good preservation of aDNA at PT1.1. Rapid desiccation, low temperatures and high salt concentration conditions at this site would have contributed to the destruction or inactivation of nucleases (Hofreiter et al., Reference Hofreiter, Serre, Poinar, Kuch and Pääbo2001), thus reducing DNA fragmentation and chemical modifications that could prevent PCR amplification. BLASTN analysis showed 99% identity with cytb sequences of P. concolor from South America and from ancient Patagonian isolates (Petrigh and Fugassa, Reference Petrigh and Fugassa2017) and 98% identity with North American puma sequences. Comparison with cytb sequences of other felids that lived in the Pleistocene showed 93 and 90% identity with Panthera onca (AN: KX419617) and 90% identity with Smilodon populator (AN: MF871700). The cytb sequence of Pleistocene P. concolor was submitted to the NCBI GenBank [Accession Number (AN): MH036361, https://www.ncbi.nlm.nih.gov/nuccore/MH036361.1] (see Appendix). Paleontological records of P. concolor have been reported in Argentina dating back to the Ensenadean (1.8–0.5 Ma) (Prevosti and Forasiepi, Reference Prevosti and Forasiepi2018). It is the only large felid surviving into the Holocene, and is still found in the region today (Mondini, Reference Mondini2002). aDNA reports from mammal coprolites before the Holocene are scarce. Most reports come from North America and date to 29 000–13 000 yr BP (e.g. Gilbert et al., Reference Gilbert, Jenkins, Götherstrom, Naveran, Sanchez, Hofreiter and Parr2008). Therefore, the molecular identification of this carnivore coprolite represents the most ancient case of South America and one of the most ancient instances in the world.

Fig. 1. Carnivore coprolite from Peñas de las Trampas 1.1 site, Argentinean Puna. Scale bar: 20 mm.
Parasitological analysis of the coprolite showed spherical or elliptical eggs. These eggs had a translucent thick wall with three layers: an outer protein membrane, a chitinous inner membrane and a thick and translucent inner layer, poorly organized, compatible with a lipid membrane. In some cases, the eggs were morulated and in others, a larva filled the inside (Fig. 2). The size of the 64 eggs was variable: 80–98.7 [92.8 ± 4.7; n = 20] × 70 – 87.5 [78.1 ± 4; n = 20] μm. This could be ascribed to the different developmental stages of the eggs. According to size and morphological characters, the eggs were similar to Toxascaris leonina. At present, this parasite has a global distribution and its definitive hosts include canids and felids, both domestic and wild (Okulewicz et al., Reference Okulewicz, Perec-Matysiak, Buńkowska and Hildebrand2012). Variable measures have been reported of T. leonina eggs, from 85 × 75 µm (Thienpont et al., Reference Thienpont, Rochette and Vanparijs1979) to 77–101 × 63–81 µm (Miyazaki, Reference Miyazaki1991). Tekele (Reference Tekele2003) measured 50 eggs and found a morphometry similar to that presented in this work: 79.06–99.07 (88.04) × 64.33–75.97 (70.12) μm. Similar eggs were found in a Middle Holocene canid coprolite from Cerro Casa de Piedra 7 archaeological site, Santa Cruz Province, in the Argentinean Patagonia, although of smaller dimensions [62.5–80 (73 ± 4.52, n = 41) × 50–72.5 (60.5 ± 4.76, n = 41) μm] (Fugassa et al., Reference Fugassa, Denegri, Sardella, Araújo, Guichón, Martinez, Civalero and Aschero2006). Additionally, eggs of 60–63 [62; n = 4] × 78 [78; n = 4] μm from a canid coprolite were recorded in a pre-Hispanic occupation of the Anasazi culture during the Late Holocene in North America (Gardner and Clary, Reference Gardner and Clary1987).
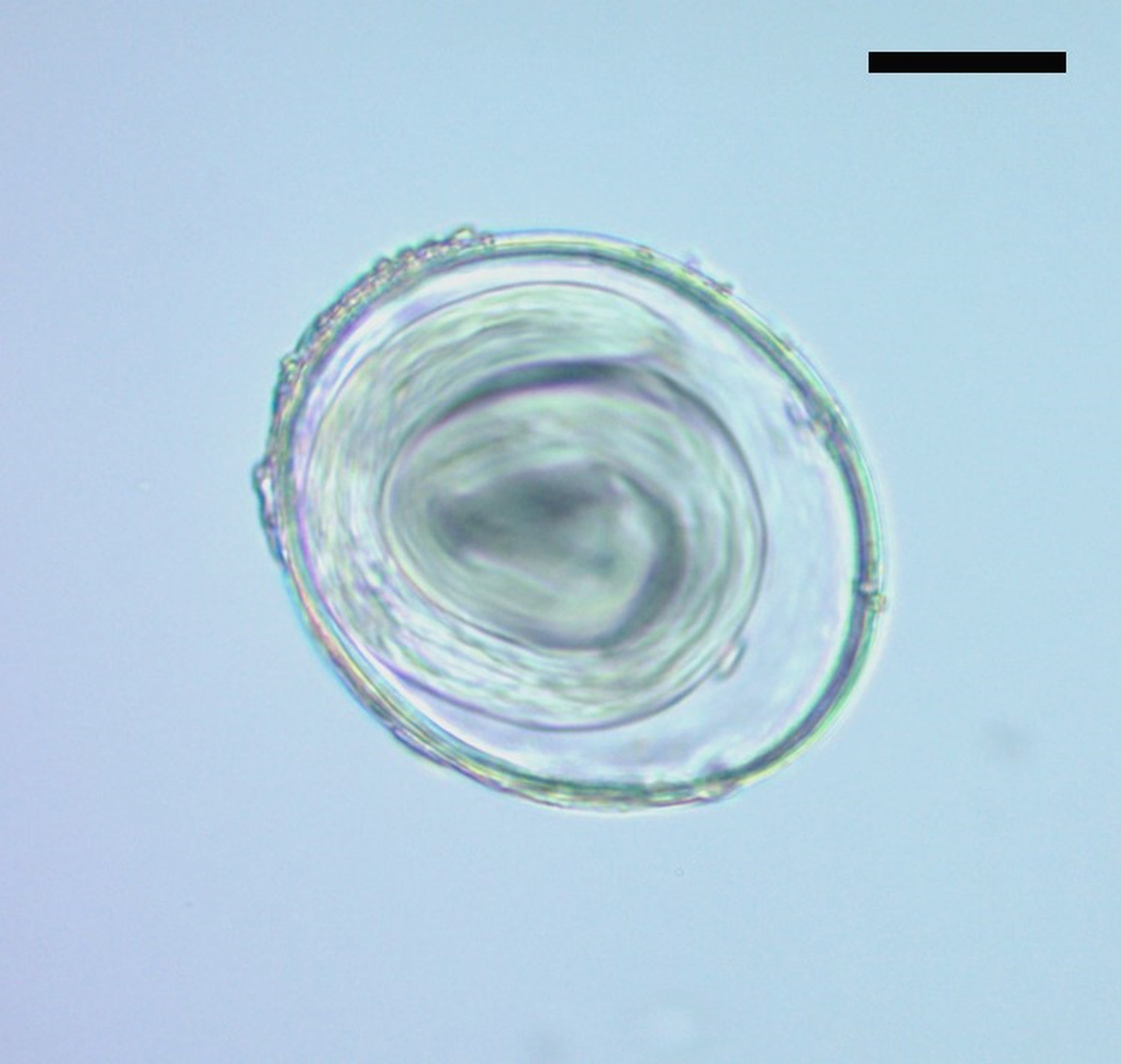
Fig. 2. Egg attributed to Toxascaris leonina (400× magnification). Scale bar: 20 µm.
A molecular approach was performed to the specific identification of eggs assigned to Toxascaris sp. A 151 bp fragment was successfully amplified in 26 replicates and no amplification was observed in two PCR replicates. An informative sequence of cox1 fragment was obtained with three positive replicates. DNA of modern T. leonina was never amplified in our laboratory. Thus, this minimized the risk of contamination of ancient T. leonina DNA with modern DNA. Therefore, the consensus sequence was assembled and submitted to the GenBank (AN: MH036362, https://www.ncbi.nlm.nih.gov/nuccore/MH036362.1) (see Appendix). The BLASTN analysis showed an identity range of 96–93% between cox1 fragment of T. leonina European and Asia isolates from different hosts (canids including dog, wolf and European fox, and felids including Eurasian lynx and South China tiger). Pairwise analysis among available T. leonina sequences showed a wide range of identity percentages, from 98 to 93%. A maximum of 1% intra-specific divergence within Iranian isolates has been reported (Mikaeili et al., Reference Mikaeili, Mirhendi, Mohebali, Hosseini, Sharbatkhori, Zarei and Kia2015). Thus, this 4% divergence with ancient T. leonina could be attributed to different temporal and geographic origins. BLASTN analysis with other ascaridid sequences showed 94–93% identity with other members of subfamily Ascaridinae: Baylisascaris spp. and Ascaris spp.
Discussion
In the present work, morphological and molecular identification of coprolite host and parasites as well as coprolite radiocarbon dating were combined to obtain accurate information on the biogeographic history of T. leonina and its host the puma, thus providing new insights into the natural history of the southern Puna.
Felids could be co-infected with Toxocara cati and T. leonina and, rarely, with Toxocara canis. The eggs of these three species are similar in their sub-spherical shape and dimension, but colour and eggshell bear differences (Okulewicz et al., Reference Okulewicz, Perec-Matysiak, Buńkowska and Hildebrand2012). It is relatively easy to differentiate Toxocara from Toxascaris eggs in modern samples. On the other hand, the comparison of DNA sequences with closely related species such as Baylisascaris spp. and Ascaris spp. showed similar identities, but eggs isolated from the puma coprolite are larger. Taphonomic conditions may alter size, shape and other characteristics of eggs (Rácz et al., Reference Rácz, Pucu, Jensen, Mostek, Morrow, van Hove, Bianucci, Willems, Heller, Araújo and Reinhard2015), but host occurrence supports egg identification because T. leonina is more commonly found in current and ancient wild felids than Baylisascaris spp. and Ascaris spp. (e.g. Acosta et al., Reference Acosta, Tantaleán and Serrano-Martínez2015; Gardner and Clary, Reference Gardner and Clary1987).
Molecular tools helped identify these ascaridid eggs, confirming the determination by morphometric characters. Records of parasite aDNA have been obtained from latrine sediments, human burials, mummy feces and, to a lesser extent, coprolites, all of them mostly human. Samples range from the Middle Holocene to historical times. On the other hand, Wood et al. (Reference Wood, Wilmshurst, Rawlence, Bonner, Worthy, Kinsella and Cooper2013) carried out an aDNA and microscopic analysis of parasites from coprolites of extinct New Zealand moa birds. The present work represents the oldest record of an aDNA sequence for a gastrointestinal nematode parasite of wild mammals, and in fact the oldest molecular parasite record worldwide, and also a new maximum age for the recovery of old DNA of this origin. Coprolite dating is unusual and ancient studies assume the age of coprolite according to that of the stratum containing it. Here, instead, coprolites themselves were dated, thus enabling a more accurate assessment of their age. This allowed confirming the presence of T. leonina in prehistoric times, presumably even before that of humans in the region, and it represents the oldest record in the world. Therefore, the common interpretation that the presence of T. leonina in modern American wild carnivores is a consequence of their contact with domestic dogs or cats (Okulewicz et al., Reference Okulewicz, Perec-Matysiak, Buńkowska and Hildebrand2012) should no longer be assumed as the only possible explanation. Canids and felids are infected by ingesting rodents and paratenic hosts and also directly by contact with feces with eggs containing infective larvae. The large number of eggs of T. leonina and its larva state in the puma coprolite analysed here indicates the high infective capacity of this parasite, involving a high risk for intermediate or paratenic hosts – including humans – and definitive hosts.
On the other hand, the authenticity of puma and T. leonina sequences was supported with the number of positive replicates and no amplification in DNA extraction and PCR-negative controls.
At a regional level, these aDNA studies have also allowed confirming the presence of pumas in the southern Puna at the end of the Pleistocene. This has significant implications for the natural history of the region, as well as for inferring the ecological context immediately previous – as far as is known so far – to the first human explorers who ventured into the area ca. 11 000 years ago (Martínez, Reference Martínez and Elizabeth2014).
Author ORCIDs
Romina S. Petrigh, 0000-0002-5016-4113
Acknowledgements
We thank Carolina Mosconi for the English edition of the manuscript, as well as Parasitology editors and referees.
Financial support
Financial support was granted by CONICET (PIP-0436, PIP-0222), FONCyT (PICT-3664), UNMdP (EXA-777) and UNC (SeCyT 30720150100108CB).
Conflict of interest
None.
Ethical standards
Not applicable.
Appendix
Nucleotide sequences
Puma concolor isolate M760 cytochrome b (cytb) gene, partial cds; mitochondrial
GenBank: MH036361.1
>MH036361.1 Puma concolor isolate M760 cytochrome b (cytb) gene, partial cds; mitochondrial
TTCTTTATCTGCCTATACATGCACGTGGGACGAGGAATATATTATGGCTCCTATACCTTCTCGGAAACATGAAATATTGGAATCATACTACTATTTGCAGTCATAGCCACAGCCTTTATAGGATATGTCTTACCATGAGGACAAATATCATTCTGAGGGGCTGCAGTT
Toxascaris leonina isolate M760 cytochrome c oxidase subunit I (COI) gene, partial cds; mitochondrial
GenBank: MH036362.1
>MH036362.1 Toxascaris leonina isolate M760 cytochrome c oxidase subunit I (COI) gene, partial cds; mitochondrial
TTTTTTGGTCATCCTGAGGTTTATATTTTAATTCTTCCTGCTTTTGGTATTATTAGTCAGAGTAGGTTGTATTTAACTGGTAAGAAAGAGGTTTTTGGTTCTTTGGGTATGGTTTATGCTATTTTGAGTATTGGTTTGATTGGTTGTGTT





