Introduction
Seeds enable plants to move through both time and space, determining where and when plants are established (Koornneef et al., Reference Koornneef, Bentsink and Hilhorst2002). The seed-to-seedling transition is therefore one of the major shifts in the plant life cycle (Finch-Savage and Leubner-Metzger, Reference Finch-Savage and Leubner-Metzger2006; Springthorpe and Penfield, Reference Springthorpe and Penfield2015).
Central to the control of the timing of the induction of germination is the role of the antagonistically acting hormones abscisic acid (ABA) and gibberellins (GA), which inhibit and promote this transition, respectively (Finkelstein et al., Reference Finkelstein, Reeves, Ariizumi and Steber2008; Holdsworth et al., Reference Holdsworth, Bentsink and Soppe2008). The hormone balance theory which describes this relationship provides a molecular thresholding mechanism by which the development state of seeds is defined (Karssen and Lacka, Reference Karssen, Lacka and Bopp1986). The most abundant hormone is thought to define whether or not a seed transitions to the germination programme (Bradford and Trewavas, Reference Bradford and Trewavas1994). Biosynthetic (Olszewski et al., Reference Olszewski, Sun and Gubler2002; Seo and Koshiba, Reference Seo and Koshiba2002) and signalling pathways for ABA (Park et al., Reference Park, Fung, Nishimura, Jensen, Fujii, Zhao, Lumba, Santiago, Rodrigues and Chow2009) and GA (Lee et al., Reference Lee, Cheng, King, Wang, He, Hussain, Lo, Harberd and Peng2002; Murase et al., Reference Murase, Hirano, Sun and Hakoshima2008) have been identified, enabling the mechanistic regulation of the molecular agents which control dormancy and germination to be investigated.
Following the decision to germinate, the embryo within a seed commences growth (Koornneef et al., Reference Koornneef, Bentsink and Hilhorst2002). This transition into a seedling is principally driven by cell expansion, rather than cell division (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014; Sliwinska et al., Reference Sliwinska, Bassel and Bewley2009). This discrete induction of growth following the initiation of the germination programme is promoted by GA (Groot and Karssen, Reference Groot and Karssen1987; Koornneef and Van der Veen, Reference Koornneef and Van der Veen1980) and its induction of gene expression associated with cell wall remodelling proteins which facilitate cell growth (Nakabayashi et al., Reference Nakabayashi, Okamoto, Koshiba, Kamiya and Nambara2005; Dekkers et al., Reference Dekkers, Pearce, van Bolderen-Veldkamp, Marshall, Widera, Gilbert, Drost, Bassel, Muller and King2013; Narsai et al., Reference Narsai, Gouil, Secco, Srivastava, Karpievitch, Liew, Lister, Lewsey and Whelan2017). These cell expansion-associated genes may be considered the downstream targets of the germination process in light of the central role they play in the regulation of embryo growth (Bassel, Reference Bassel2016).
Spatially distinct domains of gene expression programmes have been identified within the germinating Arabidopsis embryo using both gene expression analysis (Dekkers et al., Reference Dekkers, Pearce, van Bolderen-Veldkamp, Marshall, Widera, Gilbert, Drost, Bassel, Muller and King2013) and the microscopic analysis of specific reporter constructs (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014). The cellular sites of ABA and GA response and metabolism (Topham et al., Reference Topham, Taylor, Yan, Nambara, Johnston and Bassel2017), and growth-promoting cell wall-associated gene expression (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014) have also been defined at single-cell resolution. In non-germinating Arabidopsis embryos, the radicle was found to be enriched for both ABA- and GA-associated synthesis and response components, leading to the proposal that this subdomain of the embryo acts as a decision-making centre in the control of seed dormancy (Topham et al., Reference Topham, Taylor, Yan, Nambara, Johnston and Bassel2017). Examination of gene expression associated with cell wall-associated gene expression revealed this to be first induced within the embryo radicle (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014). These results collectively suggest that the radicle is where germination is initiated, a spatial site that overlaps with the decision-making centre.
The germination of seeds from an individual mother plant is typically non-uniform, with bet-hedging strategies being implemented (Bradford, Reference Bradford2002; Rowse and Finch-Savage, Reference Rowse and Finch-Savage2003; Springthorpe and Penfield, Reference Springthorpe and Penfield2015; Mitchell et al., Reference Mitchell, Johnston and Bassel2016). This strategy is believed to improve plant fitness, while mechanisms underpinning this bet-hedging behaviour have been proposed (Johnston and Bassel, Reference Johnston and Bassel2018).
In the context of food production systems, this bet-hedging trait is not favourable. Uniformity is a key objective in field-based agriculture at all stages of these systems. In order to achieve this, the germination of seeds must be synchronous once they have been planted. This co-ordinated crop establishment leads both to the suppression of weeds, and uniformity of the final product at harvest (Finch-Savage and Bassel, Reference Finch-Savage and Bassel2015). In light of this important role for uniformity in seed behaviour, procedures have been developed by commercial seed vendors which increase this population trait (Taylor et al., Reference Taylor, Allen, Bennett, Bradford, Burris and Misra1998; Paparella et al., Reference Paparella, Araújo, Rossi, Wijayasinghe, Carbonera and Balestrazzi2015). In a process termed ‘seed priming’, seeds are held in suboptimal conditions for extended periods to repress germination (Finch-Savage et al., Reference Finch-Savage, Dent and Clark2004). Following the release of the seeds from this inhibitory treatment, the resulting germination profile of the seeds is more uniform. The mechanisms by which priming acts remain poorly understood, with protocols largely focusing on the efficacy of treatments rather than the mechanisms underpinning them. This limits the potential to enhance seed quality using these approaches.
Residual dormancy represents an obstacle to germination uniformity, and priming treatments can relieve seeds of this intrinsic block to germination. One method to eliminate dormancy in temperate seeds, including Arabidopsis, is to apply a low temperature treatment, termed ‘stratification’ (Yamauchi et al., Reference Yamauchi, Ogawa, Kuwahara, Hanada, Kamiya and Yamaguchi2004). Widespread gene expression changes are associated with this low temperature response, but it remains unclear where these are located within the context of the multi-cellular embryo.
In this study we examined the spatiotemporal gene expression events underpinning the seed-to-seedling transition in Arabidopsis. We then compared these dynamic changes with those that occur in a seed that has been subjected to dormancy-relieving low temperature, in an effort to provide insight into the mechanism by which this germination-enhancing treatment is acting within seeds.
Materials and methods
Plant growth conditions
All plants in this study are in the Columbia background of Arabidopsis, and were grown in environmentally controlled cabinets, using 16 h light (23°C) and 8 h dark at 22°C. Seeds were collected when plants had stopped flowering, and placed in glassine bags for 4 weeks to reduce primary dormancy. Seeds were then cleaned by passing dried plant material through a fine mesh, and seeds were collected for use in subsequent experiments.
Germination assays
Germination of Arabidopsis thaliana seeds was conducted by plating 30 seeds in triplicate. Seeds were scored for radicle emergence (germination) every 4 h until 100% germination was reached. All seeds were on sterilized with 10% bleach and germinated on ½ MS 0.8% (w/v) agar plates in a growth room conditions with a 16 h light/8 h dark photoperiod at 22°C.
Cold treatment protocol
Arabidopsis thaliana seeds were cold treated as described previously (Sano et al., Reference Sano, Kim, Onda, Nomura, Mochida, Okamoto and Seo2017). Following 3 days of incubation at 4°C in the dark, seeds were transferred to 22°C in the light for 12 h before being dried. Seeds were dried by placing seeds between two layers or filter paper for 24 h. Reimbibition of seeds was done by plating them on ½ MS 0.8% (w/v) agar plates.
Generation of reporter constructs
Reporter constructs for EXPANSIN genes were generated using 2 kb of sequence upstream of the ATG start codon for each gene as previously described (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014). Other reporter constructs come from previous publications, including XTH18 and XTH19 (Vissenberg et al., Reference Vissenberg, Oyama, Osato, Yokoyama, Verbelen and Nishitani2005), GID1A::GID1A-GUS and GID1C::GID1C-GUS translational fusions (Suzuki et al., Reference Suzuki, Park, Okubo, Kitamura, Ueguchi-Tanaka, Iuchi, Katoh, Kobayashi, Yamaguchi and Matsuoka2009), GA3ox1::GUS and GA3ox2::GUS reporters (Hu et al., Reference Hu, Mitchum, Barnaby, Ayele, Ogawa, Nam, Lai, Hanada, Alonso and Ecker2008), SCL3::GUS (Zhang et al., Reference Zhang, Ogawa, Fleet, Zentella, Hu, Heo, Lim, Kamiya, Yamaguchi and Sun2011), AAO3::AAO3-GUS and ABA2::ABA2-GUS (Seo et al., Reference Seo, Hanada, Kuwahara, Endo, Okamoto, Yamauchi, North, Marion-Poll, Sun and Koshiba2006), and RAB18::GUS (Ghassemian et al., Reference Ghassemian, Nambara, Cutler, Kawaide, Kamiya and McCourt2000).
GUS histochemical staining
Arabidopsis thaliana embryos were dissected from seeds using a scalpel and forceps using a Leica SD6 binocular microscope. Embryos were stained in 5-bromo-4-chloro-3-indolyl-beta-d-glucuronic acid, cyclohexylammonium salt (X-Gluc) solution with 0.1 M sodium phosphate buffer (pH 7.0), 0.1% Triton X-100 and 2 mM X-Gluc (Sigma). Embryos were stained at 37°C until the blue substrate became visible, or for 24 h. Samples were fixed in a 3:1 ethanol/acetic acid, 500:1 DMSO, 1% Tween 20 fixative solution for 24 h and cleared in a chloral hydrate solution until embryos were clear for imaging. Embryos were imaged using a Leica DM500 light microscope.
Results
Cold treatment of Arabidopsis seeds
The impact of cold treatment on the speed of germination in Arabidopsis seeds was investigated by imbibing seeds in the dark at 4°C for 3 days. Seeds were then transferred to 22°C in the light for 12 h before being dried, then reimbibed. This protocol was selected based on its inclusion of a low temperature treatment and increase in the speed at which Arabidopsis seeds germinate (Sano et al., Reference Sano, Kim, Onda, Nomura, Mochida, Okamoto and Seo2017). Using both the Col and Ler ecotypes, we confirmed that the speed at which germination is completed is reduced following this process (Fig. 1).
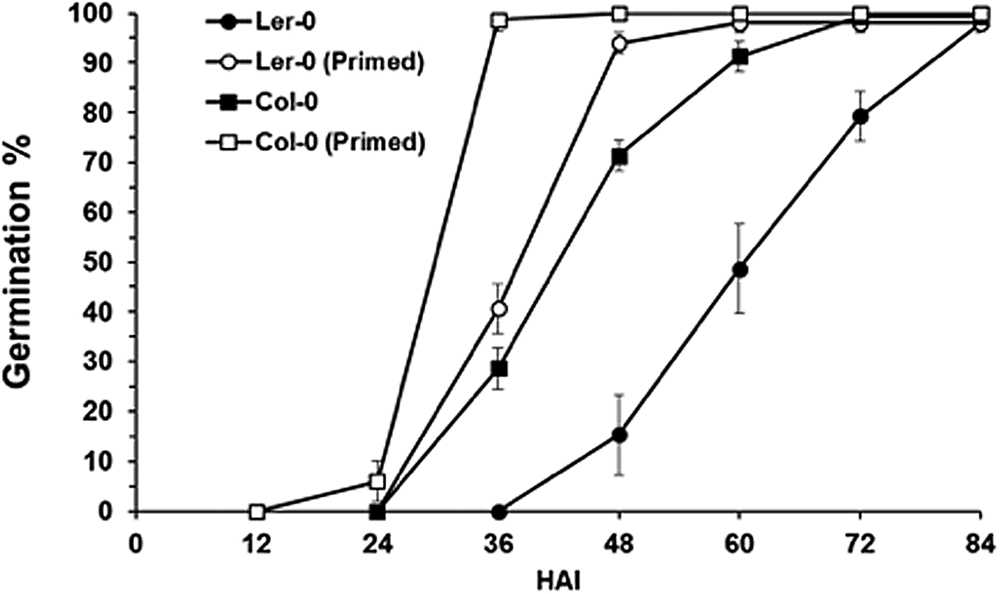
Fig. 1. Germination percentage of primed and unprimed Col-0 and Ler seeds at 22°C. Values are mean germination (%), and error bars are SE (n = 3). HAI, Hours after imbibition.
Visualizing molecular dynamics within germinating Arabidopsis embryos
Underpinning the transition from seed to seedling is a sequence of dynamic molecular events that unfold with the germinating embryo. These dynamic gene expression and epigenetic changes have been characterized previously on a genome-wide scale (Nakabayashi et al., Reference Nakabayashi, Okamoto, Koshiba, Kamiya and Nambara2005; Dekkers et al., Reference Dekkers, Pearce, van Bolderen-Veldkamp, Marshall, Widera, Gilbert, Drost, Bassel, Muller and King2013; Narsai et al., Reference Narsai, Gouil, Secco, Srivastava, Karpievitch, Liew, Lister, Lewsey and Whelan2017). We sought to build upon this work by investigating the spatiotemporal dynamics of candidate genes using reporter constructs and light microscopy. The visualization of key reporters across the developmental sequence from seed to seedling enables the dynamic processes underpinning this transition to be understood. Comparing the expression pattern of these reporters in seeds across the cold treatment process may therefore provide insight into the stage at which the treatment is arresting this dynamic developmental programme, or associated changes in the molecular programme.
Three major classes of reporter were selected. The first represent genes that encode proteins targeted to the cell wall, and promote cell expansion (Cosgrove, Reference Cosgrove2005). The other two classes represent genes and proteins associated with ABA and GA synthesis, perception and response. The optical heterogeneity of mature Arabidopsis embryos makes it not possible to visualize fluorescent proteins deep within samples (Moreno, Reference Moreno, Bougourd, Haseloff and Feijo2006). In order to achieve this, samples can be clarified and the β-glucuronidase (GUS) reporter (Jefferson, Reference Jefferson1989) observed throughout all cells of the tissue (Truernit et al., Reference Truernit, Bauby, Dubreucq, Grandjean, Runions, Barthelemy and Palauqui2008). We made use of this system to examine the spatial and temporal changes that occur in germinating Arabidopsis seeds.
GA synthesis and signalling during the seed-to-seedling transition
GA is required for the induction of germination of intact Arabidopsis seeds (Koornneef and Van der Veen, Reference Koornneef and Van der Veen1980). The patterns of gene expression associated with this induction of GA synthesis show cell type-specific profiles (Yamaguchi et al., Reference Yamaguchi, Kamiya and Sun2001; Ogawa et al., Reference Ogawa, Hanada, Yamauchi, Kuwahara, Kamiya and Yamaguchi2003), which are modulated by cold temperatures (Yamauchi et al., Reference Yamauchi, Ogawa, Kuwahara, Hanada, Kamiya and Yamaguchi2004) and light (Yamaguchi et al., Reference Yamaguchi, Kamiya and Sun2001). This in turn leads to GA responses that act to promote downstream cell wall-associated gene expression in seeds (Cao et al., Reference Cao, Cheng, Wu, Soo and Peng2006). Central to this induction are the GIBBERELLIN INSENSITIVE DWARF (GID) receptors (Ueguchi-Tanaka et al., Reference Ueguchi-Tanaka, Ashikari, Nakajima, Itoh, Katoh, Kobayashi, Chow, Yue-ie, Kitano and Yamaguchi2005), DELLA proteins (Lee et al., Reference Lee, Cheng, King, Wang, He, Hussain, Lo, Harberd and Peng2002) and the SCARECROW-LIKE3 (SCL3) transcription factor which controls germination responses (Zhang et al., Reference Zhang, Ogawa, Fleet, Zentella, Hu, Heo, Lim, Kamiya, Yamaguchi and Sun2011). GUS reporters for GA-synthesis, -signalling and -response components were examined to understand the spatiotemporal events underpinning this hormone response that stimulates the germination process.
The hormone GA is perceived by the GID1 receptors (Ueguchi-Tanaka et al., Reference Ueguchi-Tanaka, Ashikari, Nakajima, Itoh, Katoh, Kobayashi, Chow, Yue-ie, Kitano and Yamaguchi2005). The presence of GID1A or GID1C proteins is a primary requirement for a cell to be able to respond to this hormone. We examined the distribution of these receptors using GUS translational fusions (Gallego-Giraldo et al., Reference Gallego-Giraldo, Hu, Urbez, Gomez, Sun and Perez-Amador2014). During the germination process, both of these proteins were broadly distributed across the embryo, with a slight bias towards the axis (Figs 2 and 3). Following 1 day of cold treatment in the dark at 4°C, both of these proteins became enriched to the vascular cells of the embryo (Figs 2L and 3L). The localization persisted until 3 days in the cold at 4°C and in the light at 22°C for a further 12 h (Figs 2M–N and 3M–N). Dehydration and rehydration of seeds did not alter the localization of GA receptors to the vasculature (Figs 2O and 3O). Cold treatment, therefore, altered the cellular site of perception of the germination-stimulating hormone GA from across the embryo to principally within the vasculature.

Fig. 2. Spatial and temporal dynamics of the GID1A::GID1A-GUS reporter during the seed-to-seedling transition in Arabidopsis. GID1A protein abundance in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) just testa ruptured (JTR), (G) late testa rupture (STR), and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of GID1A::GID1A-GUS activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.

Fig. 3. Spatial and temporal dynamics of the GID1C::GID1C-GUS reporter during the seed-to-seedling transition in Arabidopsis. GID1C protein abundance in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) early testa rupture, (G) late testa rupture, and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of GID1C::GID1C-GUS activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.
The SCL3::GUS reporter acts as a useful proxy to understand the cellular sites where GA responses are occurring (Zhang et al., Reference Zhang, Ogawa, Fleet, Zentella, Hu, Heo, Lim, Kamiya, Yamaguchi and Sun2011). Activity of this reporter is enriched in the radicle during the early stages of seed germination (Fig. 4A–E) and progressively moves along the hypocotyl and into the cotyledons until the completion of germination (Fig. 4F–H). Following germination, SCL3::GUS activity is enriched in the root (Fig. 4I–K), and presumably in the endodermis where it has been reported previously to be present (Zhang et al., Reference Zhang, Ogawa, Fleet, Zentella, Hu, Heo, Lim, Kamiya, Yamaguchi and Sun2011). SCL3::GUS activity was enriched within the vasculature from the earliest stage of cold treatment sampled until the rehydration of seeds (Fig. 5L–O). GA response as indicated by this reporter therefore closely followed the pattern of GA perception indicated by the GID1A and GID1C reporters.

Fig. 4. Spatial and temporal dynamics of the SCL3::GUS reporter during the seed to seedling transition in Arabidopsis. Promoter activity in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) early testa rupture, (G) late testa rupture, and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of SCL3::GUS promoter activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.
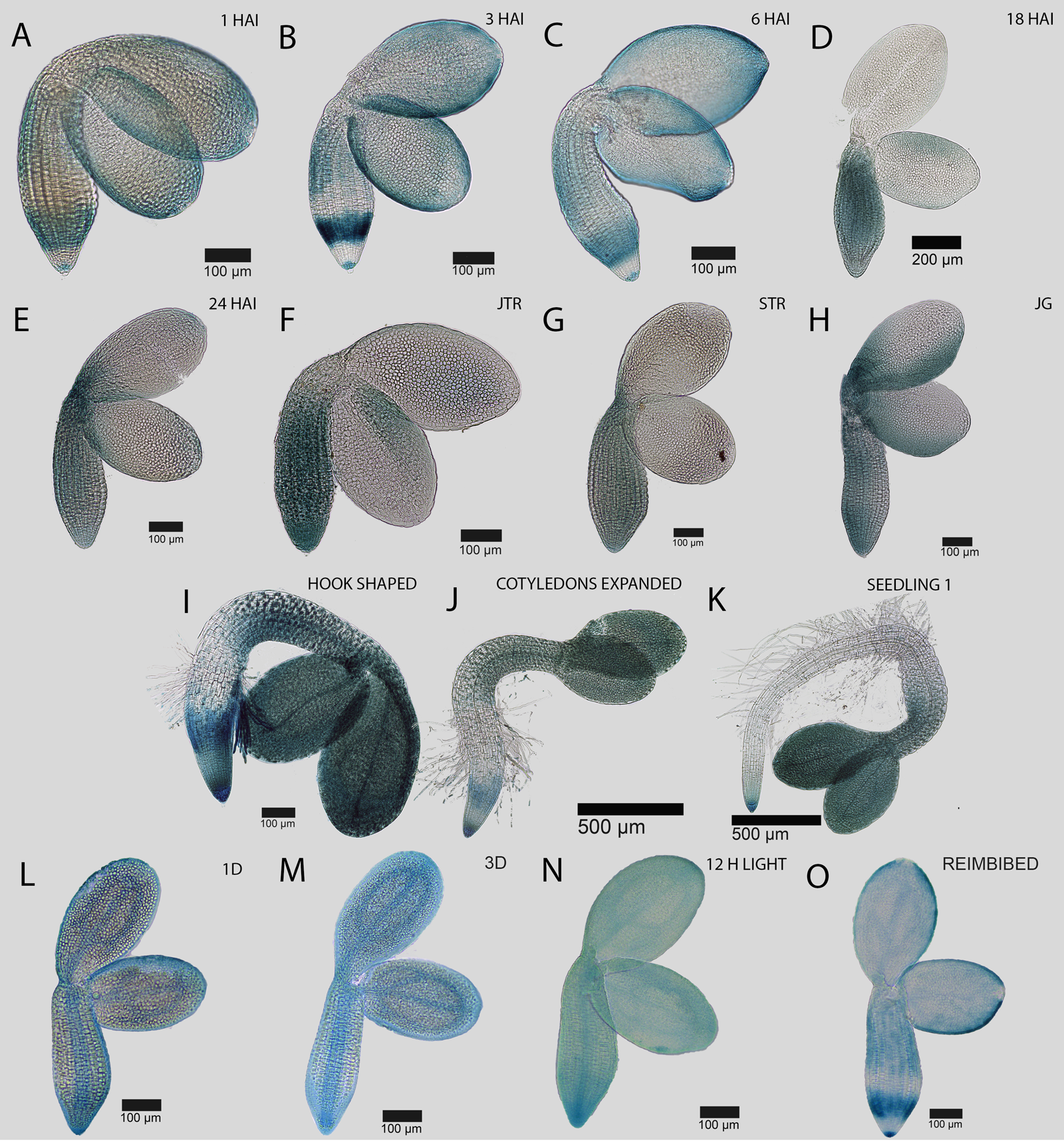
Fig. 5. Spatial and temporal dynamics of the EXPA1::GUS reporter during the seed-to-seedling transition in Arabidopsis. Promoter activity in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) early testa rupture, (G) late testa rupture, and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of EXPA1::GUS promoter activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.
The final step and rate-limiting of GA synthesis is catalysed by the enzyme GIBBERELLIN 3 β-HYDROXYLASE (GA3ox) (Olszewski et al., Reference Olszewski, Sun and Gubler2002). Examination of the GA3ox1::GUS promoter reporter (Mitchum et al., Reference Mitchum, Yamaguchi, Hanada, Kuwahara, Yoshioka, Kato, Tabata, Kamiya and Sun2006) indicated a broad initial induction across the embryo radicle, followed by movement into the hypocotyl as germination progresses (Supplementary Fig. S1A–G). In response to cold, a broad domain of activity of this promoter was observed across the embryo (Supplementary Fig. S1L–N). This GA-synthesis promoter therefore does not become enriched within the vasculature as do the perception and response components.
Cell growth-associated gene expression during the seed-to-seedling transition
Gene expression for enzymes that promote cell wall weakening act as a proxy to understand which cells are having their growth promoted (Cosgrove, Reference Cosgrove2005). These downstream targets of the germination process have been demonstrated to play a regulatory role in the control of this transition (Lü et al., Reference Lü, Kang, Jiang, Dai, Gao and Zhang2013). Proteins encoded by EXPANSIN (EXPA) and XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE (XTH) genes promote cell expansion in Arabidopsis cells (Vissenberg et al., Reference Vissenberg, Oyama, Osato, Yokoyama, Verbelen and Nishitani2005). We examined the activity of the promoters encoding members of the EXPA and XTH multigene families that are conditionally unregulated in response to the induction of the germination program (Nakabayashi et al., Reference Nakabayashi, Okamoto, Koshiba, Kamiya and Nambara2005; Bassel et al., Reference Bassel, Fung, Chow, Foong, Provart and Cutler2008; Dekkers et al., Reference Dekkers, Pearce, van Bolderen-Veldkamp, Marshall, Widera, Gilbert, Drost, Bassel, Muller and King2013).
The spatiotemporal patterns of gene expression associated with growth-promoting cell wall modifying gene expression may serve as a proxy to understand spatial control of cell expansion (Cosgrove, Reference Cosgrove2005). A series of expansin genes are induced in germinating Arabidopsis embryos (Nakabayashi et al., Reference Nakabayashi, Okamoto, Koshiba, Kamiya and Nambara2005) and promoter-GUS reporters to these genes have been generated previously (Bassel et al., Reference Bassel, Stamm, Mosca, de Reuille, Gibbs, Winter, Janka, Holdsworth and Smith2014; Stamm et al., Reference Stamm, Topham, Mukhtar, Jackson, Tomé, Beynon and Bassel2017). These lines were examined using light microscopy over the time course of seed germination and early seedling establishment to explore the genetic control of cell expansion. EXPA1::GUS is first induced in cells of the embryonic radicle, and spreads progressively up the axis and into the cotyledons during germination (Fig. 5A–F). Following germination, promoter activity is increasingly focused to the root tip and cotyledons, being excluded from the majority of the root and hypocotyl (Fig. 5G–K).
During cold treatment, the EXPA1::GUS promoter becomes enriched within the vascular cells of the embryo. This follows the pattern of upstream GA perception and response (Figs 2–4). Following the cold exposure, the spatiotemporal pattern observed between 3–6 hours after imbibition (HAI) of regular germination (Fig. 5B,C). The GA-responsive promoter EXPA1::GUS therefore follows the vasculature relocalization of gene expression associated with stratification. This is, however, not strictly true for all cell wall remodelling enzyme promoters, and was not observed for either EXPA15::GUS (Supplementary Fig. S2) or XTH19::GUS (Supplementary Fig. S3).
ABA synthesis and signalling during the seed-to-seedling transition
ABA plays a central role in the control of seed dormancy, and in mediating stress responses, limiting the germination of non-dormant seeds (Kushiro et al., Reference Kushiro, Okamoto, Nakabayashi, Yamagishi, Kitamura, Asami, Hirai, Koshiba, Kamiya and Nambara2004). We examined the expression of reporters associated with ABA synthesis and response over the time course of germination and compared this with the pattern in cold-treated seeds to determine at which stage low temperature arrests the ABA programme.
The final step of ABA synthesis is catalysed by the enzyme ALDEHYDE OXIDASE 3 (AAO3) (Seo et al., Reference Seo, Hanada, Kuwahara, Endo, Okamoto, Yamauchi, North, Marion-Poll, Sun and Koshiba2006). The distribution of this protein is principally localized to the embryo axis during germination (Fig. 6A–G). During the early stages of cold treatment, the protein becomes enriched in the vasculature (Fig. 6L), where it becomes progressively less abundant. A similar pattern of ABA synthesis protein redistribution is with the ABA-synthesis protein ABA DEFICIENT 2 (ABA2), which catalyses the penultimate step in hormone synthesis. During the early stages of germination, the protein is most abundant in the radicle (Fig. 7A–G), yet during low temperature exposure becomes enriched within the vasculature (Fig. 7L–O). The sites of ABA synthesis overlap with those of GA perception and response during and following Arabidopsis seed stratification.

Fig. 6. Spatial and temporal dynamics of the AAO3::AAO3-GUS reporter during the seed to seedling transition in Arabidopsis. AAO3 protein abundance in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) early testa rupture, (G) late testa rupture, and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of AAO3::AAO3-GUS activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.

Fig. 7. Spatial and temporal dynamics of the ABA2::ABA2-GUS reporter during the seed to seedling transition in Arabidopsis. ABA2 protein abundance in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) early testa rupture, (G) late testa rupture, and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of ABA2::ABA2-GUS activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.
The RESPONSE TO ABA18 (RAB18) promoter has been characterized previously as responsive to ABA, and acts as a useful proxy to determine where ABA-mediated transcriptional responses are occurring (Lång and Palva, Reference Lång and Palva1992). Over the early stages of germination, this reporter is principally concentrated in the radicle, with some activity present on the outer margins of the cotyledons (Fig. 8A–D). This pattern continues until the seed has completed germination (Fig. 8E–H), where it then becomes focused first in the root of the early seedling (Fig. 8J) then the lower hypocotyl at a later stage of seedling development (Fig. 8K). Embryos from seeds that are being stratified at 4°C in the dark show RAB18::GUS in the vasculature, but this then changes to a spotty pattern in the axis and cotyledons once seeds are moved to 22°C in the light (Fig. 8L–O). ABA response therefore follows a vascular localization, but only at low temperatures.

Fig. 8. Spatial and temporal dynamics of the RAB18::GUS reporter during the seed-to-seedling transition in Arabidopsis. Promoter activity in the germinating embryo at (A) 1 HAI, (B) 3 HAI, (C) 6 HAI, (D) 18 HAI, (E) 24 HAI, (F) early testa rupture, (G) late testa rupture, and germinated seedlings just after the completion of germination (H), (I) hook stage seedling, (J) recently expanded cotyledons and (K) a fully established seedling. Pattern of RAB18::GUS promoter activity in an embryo following (L) 1 day and (M) 3 days of priming treatment at 4°C in the dark, (N) 12 h at 22°C in the light, and (O) following reimbibition. Black bars indicate the scale in each image.
Discussion
This study sought to investigate how low germination-stimulating temperatures impact the spatiotemporal dynamics of GA and ABA. By characterizing the sequential steps underlying seed germination for gene expression associated with cell growth, and hormone synthesis and response, a molecular developmental chronology was established. This was compared with seeds during the low temperature treatment to determine whether spatial changes were invoked by this treatment.
Generally speaking, molecular components and promoter activities were redistributed to the vascular cells of the embryo after low temperature treatment. This was observed for GA-perception, GA-response, cell wall modifying gene expression, ABA-synthesis and ABA-response-associated genes and proteins. The reason why this redistribution occurs remains unclear; however, the ubiquity with which it happens across a broad range of reporters suggests that placing all components into a single cell type is consistent.
In primary dormant Arabidopsis seeds, the cells that respond to each ABA and GA are enriched within the radicle, and separated in the vasculature and root cap, respectively (Topham et al., Reference Topham, Taylor, Yan, Nambara, Johnston and Bassel2017). This spatial separation endowed the system with the ability to process alternating temperatures. This ability to process changing temperature was lost in models where hormone responses are contained within the same cell. The spatial relocalization of both ABA- and GA-signalling components to the vasculature following cold treatment suggests that this complex temperature processing capacity is absent. This may be a consequence to residual dormancy being alleviated and this no longer being needed. This work sheds light on the molecular and cellular basis of cold-stimulated seed germination, and may provide cellular targets for genetic manipulation to enhance seed quality.
Supplementary Material
To view Supplementary Material for this article, please visit: https://doi.org/10.1017/S0960258519000266.
Financial support
N.K.M. was funded by a scholarship of Academic Training Scheme (SLAI) from the Ministry of Education Malaysia, Faculty of Agro Based Industry, University Malaysia Kelantan, Jeli Campus, and G.W.B. was funded by BBSRC grants BB/L010232/1, BB/J017604/1 and BB/N009754/1, and Leverhulme grant RPG-2016-049.
Conflicts of interest
The authors declare no conflicts of interest.










