INTRODUCTION
Schistosomes are important blood-fluke parasites of humans and domestic livestock (Rollinson et al. Reference Rollinson, Kaukas, Johnston, Simpson and Tanaka1997). These trematodes are divided into 4 main groups: Schistosoma mansoni group, S. haematobium group, S. indicum group and S. japonicum group (Secor and Colley, Reference Secor and Colley2005). Echinostomes are also trematodes but, unlike schistosomes, they develop and are restricted to the intestinal lumen of the definitive host and do not have a tissue invasive phase (Toledo and Fried, Reference Toledo and Fried2005; Toledo et al. Reference Toledo, Esteban and Fried2009).
Human schistosomiasis is a neglected tropical disease and a major public health concern in Africa, the Middle East, Asia and South America. Some 200 million people are infected with schistosomes, with a further 700 million at risk of infection in tropical and subtropical regions (Engels et al. Reference Engels, Chitsulo, Montresor and Savioli2002). As there is currently no available vaccine for this disease in people (Bergquist et al. Reference Bergquist, Utzinger and McManus2008), the foundation of control is based upon provision of chemotherapy to afflicted communities, in particular mass drug administration of the anthelmintic praziquantel (Doenhoff et al. Reference Doenhoff, Hagan, Cioli, Southgate, Pica-Mattoccia, Botros, Coles, Tchueme-Tuchuente, Mbaye and Engels2009). However, the search for an effective vaccine continues to be a key priority (Secor and Colley, Reference Secor and Colley2005).
Urinary schistosomiasis, caused by Schistosoma haematobium, is the most prevalent form of schistosomiasis in Africa and the Middle East. Children carry the heaviest burden of infection with as many as 100% of primary school children infected in areas such as our study sites in Zimbabwe (Midzi et al. Reference Midzi, Sangweme, Zinyowera, Mapingure, Brouwer, Munatsi, Mutapi, Mudzori, Kumar, Woelk and Mduluza2008). Children younger than school-age can also be infected and begin to exhibit disease (Garba et al. Reference Garba, Barkier, Djibo, Lamine, Sofo, Gouras, Bosque-Oliva, Webster, Stothard, Utzinger and Fenwick2010). As a result, schistosome-related morbidities include both non-immunological forms (blood in the urine, pain during urination, anaemia, growth retardation, poor cognition and memory) and immune-mediated forms (tissue damage and organomegaly) (Midzi et al. Reference Midzi, Sangweme, Zinyowera, Mapingure, Brouwer, Munatsi, Mutapi, Mudzori, Kumar, Woelk and Mduluza2008). Immuno-pathological reactions against schistosome eggs trapped in the tissues leads to inflammatory and obstructive disease in the bladder, ureter and kidney as well as fibrosis. Urinary schistosomiasis likely predisposes to bladder cancer and HIV infection (Stoever et al. Reference Stoever, Molyneux, Hotez and Fenwick2009).
To develop vaccines protective against infection and/or pathology based on natural immune responses against schistosomes, there is ongoing research both in humans and animals in a context of experimental and natural schistosomiasis (Hagan et al. Reference Hagan, Blumenthal, Dunne, Simpson and Wilkins1991; Dunne et al. Reference Dunne, Butterworth, Fulford, Ouma and Sturrock1992; Demeure et al. Reference Demeure, Rihet, Abel, Ouattara, Bourgois and Dessein1993; Grogan et al. Reference Grogan, Kremsner, Van Dam, Deelder and Yazdanbakhsh1997; Mutapi et al. Reference Mutapi, Ndlovu, Hagan, Spicer, Mduluza, Turner, Chandiwana and Woolhouse1998). Several studies have demonstrated similarities between different Schistosoma species in terms of life-histories and immunological aspects (Verjovski-Almeida et al. Reference Verjovski-Almeida, DeMarco, Martins, Guimaraes, Ojopi, Paquola, Piazza, Nishiyama, Kitajima, Adamson, Ashton, Bonaldo, Coulson, Dillon, Farias, Gregorio, Ho, Leite, Malaquias, Marques, Miyasato, Nascimento, Ohlweiler, Reis, Ribeiro, Sa, Stukart, Soares, Gargioni, Kawano, Rodrigues, Madeira, Wilson, Menck, Setubal, Leite and Dias-Neto2003; Capron et al. Reference Capron, Riveau, Capron and Trottein2005; Berriman et al. Reference Berriman, Haas, LoVerde, Wilson, Dillon, Cerqueira, Mashiyama, Al-Lazikani, Andrade, Ashton, Aslett, Bartholomeu, Blandin, Caffrey, Coghlan, Coulson, Day, Delcher, DeMarco, Djikeng, Eyre, Gamble, Ghedin, Gu, Hertz-Fowler, Hirai, Hirai, Houston, Ivens, Johnston, Lacerda, Macedo, McVeigh, Ning, Oliveira, Overington, Parkhill, Pertea, Pierce, Protasio, Quail, Rajandream, Rogers, Sajid, Salzberg, Stanke, Tivey, White, Williams, Wortman, Wu, Zamanian, Zerlotini, Fraser-Liggett, Barrell and El-Sayed2009; Zhou et al. Reference Zhou, Zheng, Chen, Zhang, Wang, Guo, Huang, Zhang, Huang, Jin, Dou, Hasegawa, Wang, Zhang, Zhou, Tao, Cao, Li, Vinar, Brejova, Brown, Li, Miller, Blair, Zhong, Chen, Liu, Hu, Wang, Zhang, Song, Chen, Xu, Xu, Ju, Huang, Brindley, McManus, Feng, Han, Lu, Ren, Wang, Gu, Kang, Chen, Chen, Chen, Wang, Yan, Wang, Lv, Jin, Wang, Pu, Zhang, Zhang, Hu, Zhu, Wang, Yu, Wang, Yang, Ning, Beriman, Wei, Ruan, Zhao, Wang, Liu, Zhou, Wang, Lu, Zheng, Brindley, McManus, Blair, Zhang, Zhong, Wang, Han, Chen, Wang, Han and Chen2009), but little is known about molecular phenotypic differences that may be involved in host adaptation which might affect the efficacy of future vaccines. Despite the demonstration that antibody-mediated responses can protect against schistosome infection in experimental models, current human schistosome vaccine research, based on antibody-mediated protection, has stalled with the failure of many of the vaccine candidate antigens to enter Phase III clinical trials (Hagan and Sharaf, Reference Hagan and Sharaf2003). Limitations in our current understanding of the development of protective anti-schistosome responses against specific antigenic proteins as well as the parasite's biology (particularly antigen expression patterns) may be contributing to the slow development of effective anti-schistosome vaccines.
To shed light on these issues, comparison of the protein expression of S. haematobium adult worms with other trematode parasites could be illuminating especially in reference to S. bovis which is a ‘molecular’ analogue of S. haematobium and an experimental model for vaccine research (Capron et al. Reference Capron, Riveau, Capron and Trottein2005). Comparison with other more distantly-related trematodes, e.g. Echinostoma caproni, is also useful by providing inferences into putative responses to different life history tracts, i.e. echinostomes do not have a tissue phase in the definitive host, and present an opportunity to investigate host-related adaptations in protein expression patterns. Although S. bovis and S. haematobium differ in their definitive hosts and in their niches within the host vasculature (Vercruysse and Gabriel, Reference Vercruysse and Gabriel2005), being sufficiently closely related in terms of evolutionary distance (Bowles et al. Reference Bowles, Blair and McManus1995; Webster et al. Reference Webster, Southgate and Littlewood2006), they have an ability to hybridise (Huyse et al. Reference Huyse, Webster, Geldof, Stothard, Diaw, Polman and Rollinson2009). As S. bovis is much easier to keep in laboratory passage in rodents (Agnew et al. Reference Agnew, Murare, Lucas and Doenhoff1989) as well as eliciting similar cross-immunogenic profiles (Losada et al. Reference Losada, Chacón, Colmenares, Bermúdez, Lorenzo, Pointier, Theron, Alarcón de Noya and Noya2005), makes study of S. bovis particularly informative. However, significant differences are known: for example, early studies of the S. haematobium vaccine candidate glutathione-S-transferase (28 kDa GST) showed inter-species variation in the coding regions of S. haematobium vs. S. bovis vs. S. japonicum 28 kDa GST. This variation gives rise to phenotypic differences associated with host immunity (Trottein et al. Reference Trottein, Godin, Pierce, Sellin, Taylor, Gorillot, Silva, Lecocq and Capron1992).
To date, several studies using proteomic approaches have compared protein expression patterns between different helminth life stages (Curwen et al. Reference Curwen, Ashton, Johnston and Wilson2004; Jolly et al. Reference Jolly, Chin, Miller, Bahgat, Lim, DeRisi and McKerrow2007; Wang et al. Reference Wang, Abubucker, Martin, Wilson, Hawdon and Mitreva2010), including parasites of different sexes and parasite development in different hosts (Toledo et al. Reference Toledo, Espert, Munoz-Antoli, Fried and Esteban2004; Cheng et al. Reference Cheng, Lin, Feng, Fu, Jin, Yuan, Zhou and Cai2005). There have been no comparative proteomic studies on different trematode species which could lead to novel intervention targets with broader spectra and a better understanding of parasite-related host immune modulation (Harnett and Harnett, Reference Harnett and Harnett2010). Previous evolutionary and ecological studies have been carried out using genetic techniques such as micro-array (transcriptome) or genome sequencing (Cieslak and Ribera, Reference Cieslak and Ribera2009) and these have given important insights into the biology of the parasites. These techniques do not take into account post-transcriptional regulation of protein expression (López, Reference López2007; Schrimpf and Hengartner, Reference Schrimpf and Hengartner2010) and cannot determine the degree of epitope cross-reactivity between parasite species. Moreover, the proteomic approach is particularly useful in non-model organisms (López, Reference López2007; Ramm et al. Reference Ramm, McDonald, Hurst, Beynon and Stockley2009). Comparative proteomic approaches have been successfully used in other more general molecular studies: for example, assessing the divergence between different rodent species (Aquadro and Avise, Reference Aquadro and Avise1981).
In this study, we have used a proteomic approach to compare phenotypic differences between the three different parasite species in terms of protein expression and immunogenicity. We compared protein expression patterns and immune cross-reactivity between S. haematobium, S. bovis and E. caproni which may indicate proteins involved in the adaptation to different hosts and different niches potentially warranting further scrutiny as potential vaccines targets for schistosomiasis as well as several other trematode diseases.
MATERIALS AND METHODS
Parasites and experimental infections
The techniques used for the maintenance of Echinosotoma caproni in the laboratory have been described in detail elsewhere (Toledo et al. Reference Toledo, Espert, Munoz-Antoli, Fried and Esteban2004). Briefly, encysted metacercariae of E. caproni were removed from the kidneys and pericardial cavities of experimentally infected Biomphalaria glabrata snails and used to infect golden hamsters (Mesocricetus auratus). Outbred male golden hamsters, weighing 45–60 g, were infected through a stomach tube with 75 metacercariae each of E. caproni. The worm egg release by each animal was monitored daily as described previously (Toledo et al. Reference Toledo, Espert, Carpena, Muñoz-Antoli and Esteban2003). Soluble adult worm antigens (SWAP) were prepared from adult worms collected from the intestine of hamsters 6 weeks post-infection with 100 metacercariae of E. caproni following previously published protocols (Toledo et al. 2003). For S. haematobium infections used for the serological studies, parasite eggs obtained from urine of S. haematobium-infected children in Zanzibar (Stothard et al. Reference Stothard, Mgeni, Khamis, Seto, Ramsan and Rollinson2002) were hatched and used to infect Bulinus wrighti snails with 5 miracidia per snail. Upon infection patency 150 cercariae were pooled from these shedding snails and used to infect golden hamsters by the paddling technique; all experiments were in accordance with ethical principles in animal research and Home Office (UK) approvals.
Adult S. haematobium SWAP was obtained freeze dried from the Theodor Bilharz Institute (Giza, Egypt). To prepare this fraction, worms were perfused in saline buffer from hamsters, washed in PBS (pH 7·4), homogenized, centrifuged to obtain the soluble fraction and freeze-dried in aliquots (5 mg/mL). These were reconstituted with distilled water as required. Freeze-dried adult S. bovis SWAP from sheep was prepared as previously described in detail elsewhere (Oleaga and Ramajo, Reference Oleaga and Ramajo2004). SWAP preparations were prepared following similar protocols to reduce proteome variations due to different preparation approaches.
Rodent sera
For the immunological cross-reactivity assays, the antigen recognition patterns of sera from hamsters infected with S. haematobium and E. caproni were determined. For E. caproni, a pool was made from sera collected at 5, 6 and 10 weeks post-infection (hamsters normally make parasite-specific antibodies from 5 weeks) from 5 hamsters. After clotting overnight at 4°C, serum was separated from the clot by centrifugation. All the sera and the antigens were stored at −20°C until use. For sera from schistosome infected hamsters, Syrian golden hamsters were infected with 150 cercariae by paddling and bled 12 weeks post-infection. After clotting, blood collected from each hamster was centrifuged at 1400 g for 5 min to collect sera which were snap frozen in liquid nitrogen for long-term storage in liquid nitrogen. A pool of sera was made from 5 hamsters for use in this study. There were no experiments of hamsters infected with S. bovis parasites.
Preparations for CyDye labelling for DIGE
CyDye DIGE Fluor minimal dyes (GE Healthcare) were reconstituted following the manufacturer's instructions. 50μg protein of each sample were labelled with either Cy3 or Cy5. The sample volumes were adjusted to 18μL with labelling buffer (7M urea, 2M thiourea, 4% CHAPS, (w/v), 25 mM Tris Base; pH 8·5), followed by addition of 1μL dye (400 pmol) to each individual sample. The samples were left on ice for 30 minutes in the dark, followed by adding 1μL of 10 mmol/L lysine to stop the reaction.
Two-dimensional differential in gel electrophoresis (2D-DIGE)
To compare the parasite proteomes in 2D-DIGE assays, three gels were ran – one for each pair of samples. Differentially labelled samples were mixed into the same tube with 210μl of rehydration buffer (7M urea, 2M thiourea, 4% CHAPS, 5% DTE (dithioerythritol), 0·8% IPG buffer 3–10 pH and bromophenol blue). Thereafter, the first dimension i.e. isoelectric focusing (IEF) and second dimension were run following previously described protocols (Mutapi et al. Reference Mutapi, Burchmore, Mduluza, Foucher, Harcus, Nicoll, Midzi, Turner and Maizels2005) using the IEF protocol for 13 cm IPG strips; rehydration for 14 h at 20 V, 500 V for 1 h, 1000 V for 1 h and 8000 V for 3 h and performing the second dimension using 12% polyacrylamide gels with SDS buffer. Images from these gels were subsequently analysed as described below.
Image analysis and mass spectrometry
Gels were scanned on a Typhoon spectrophotometer (GE Healthcare) at the appropriate excitation/emission wavelength for each fluorophore Cy3 (532/580 nm) and Cy5 (633/670 nm) at 50 microns resolution. The images were analyzed using the Difference In–gel Analysis (DIA) module of Decyder software version 7.0 (GE Healthcare). The protein spots showing greater than 5-fold differences in relative abundance between parasite preparations were considered as differentially expressed proteins. The 5-fold difference was used to reduce the likelihood of detecting spurious differences. Proteins from the different trematodes were identified by comparing DIGE images with the proteomic map of S. haematobium (Mutapi et al. Reference Mutapi, Burchmore, Mduluza, Foucher, Harcus, Nicoll, Midzi, Turner and Maizels2005) and E. caproni (Sotillo et al. Reference Sotillo, Valero, Sánchez Del Pino, Fried, Esteban, Marcilla and Toledo2010) since there is no complete genome or protein sequence available for any of the three species studied. S. haematobium protein identities on the proteome map were obtained from a Coomassie Blue-stained reference gel which had been prepared and processed to obtain MS/MDS data which were submitted for an MS/MS ion search via the Mascot search engine (Matrix Science), and non-redundant National Center for Biotechnology Information (NCBI) database (Mutapi et al. Reference Mutapi, Burchmore, Mduluza, Foucher, Harcus, Nicoll, Midzi, Turner and Maizels2005). Briefly, plugs of 1·4 mm were excised from the reference Coomassie Blue-stained gel and subjected to in-gel trypsin digestion in an Ettan Spot Handing Workstation (GE Healthcare), in accordance with standard protocols (Amersham).
The resulting tryptic peptides were solubilized in 0·5% formic acid and were fractionated by nanoflow high-performance liquid chromatography on a C18 reverse phase column (GE Healthcare), and elution was performed with a continuous linear gradient of 40% acetonitrile for 20 min. The elutants were analyzed by online electrospray tandem MS (MS/MS) by use of a Qstar Pulsar mass spectrometer (Applied Biosystems). A 3 sec survey scan preceded each MS/MS data-collection cycle of 4 product ion scans of 3 sec each, and this gave a duty cycle of 15 sec. Data were submitted for an MS/MS ion search via the Mascot search engine (Matrix Science), and both locally established databases for S. mansoni EST sequences and the present non-redundant National Center for Biotechnology Information (NCBI) database were searched.
Two-dimensional electrophoresis and Western blotting
In order to determine cross-reactive antigens, 2D gel electrophoresis (2DE) was conducted on 7 cm gels as above, with some modifications. 100 μg of protein were solubilised in rehydration buffer (7 M urea, 2 M thiourea, 4% CHAPS (w/v), 65 mM DTE and trace bromophenol blue) and 0·8% IPG buffer (pH 3–10) to make a total volume of 125μL. Each protein preparation was then added to a 7 cm linear pH 3–10 IPG strip and the IEF was performed following the following protocol (1) passive rehydration for 14 h ; (2) 500 V for 30 min; (3) 1000 V for 30 min; (4) 8000 V for 4 h followed by equilibration in 2 mL of 1% DTE for 15 min and 2 mL of 4% iodoacetamide in equilibration buffer containing 6 M urea, 0·375 M Tris pH 8·8, 2% SDS and 20% glycerol. The second dimension was performed using 10% polyacrylamide precast gels from Invitrogen. Proteins from SDS–PAGE were stained with Coomassie blue or transferred onto nitrocellulose membranes in 25 ml 20X transfer buffer (Invitrogen), methanol 10% (v/v). After confirming transfer by staining with 0·1% Ponceau S (Sigma), membranes were blocked with TBS Start Block buffer T20 (Invitrogen) for 1 h at room temperature. After washing with TBS containing 0·05% Tween-20 (TBST), blots were incubated overnight at 4°C with a pool of 10 serum samples of E. caproni-infected hamsters, or S. haematobium-infected hamsters or negative control sera at 1:200 dilution in TBS Start Block buffer. The membrane was then washed three times for 10 min each time in TBS, 0·05% Tween 20, 0·5% Triton-X100 (TBS/TT). Bound antibodies were detected by incubating blots for 1 h at RT with horseradish peroxidase (HRP)-conjugated rabbit anti-Syrian hamster IgG (Abcam), in blocking buffer. After washing four times for 10 min each time in TBS/TT and once in TBS alone, recognised antigens were visualized using ECL Plus (Amersham) following the manufacturer's instructions, and exposed to X-OMAT film (Kodak) for 10 sec. Images from Western blotting and Coomassie blue staining were digitalised and matched by using ImageMaster software (GE Healthcare).
RESULTS
Proteome comparisons
The 2D-DIGE gels were run comparing the 3 proteomes as shown in Fig.1. DIA analysis of the gels showed both quantitative and qualitative differences. There was more similarity between the two schistosome species than between Echinostoma and Schistosoma. On the first gel comparing S. haematobium and S. bovis, 1701 spots representing different proteins (including different isoforms) were detected, with 91% showing similar expression levels (Fig. 2A). 5·4% of the proteins showed increased expression in S. haematobium by our criteria of 5-fold or greater difference in abundance on the gel while 3·6% showed increased expression in S. bovis. On the second gel, comparing S. haematobium vs. E. caproni, 1967 spots were detected with 81% showing similar expression levels. 8·4% of the protein spots showed increased expression in S. haematobium and 10·6% showed increased expression in E. caproni while 81% were present in similar amounts on both gels (Fig. 2B).
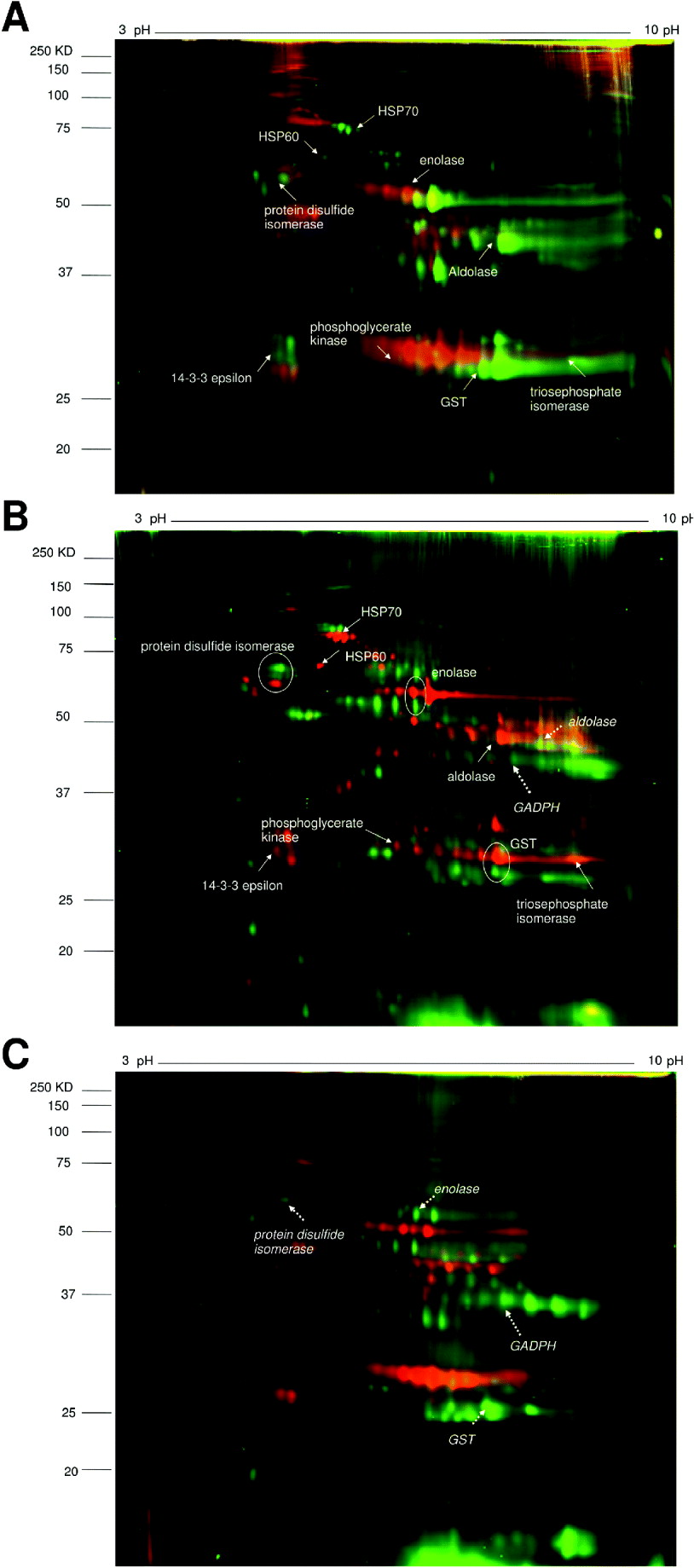
Fig. 1. 2D-DIGE images comparing pairs of different helminth species’ adult worm proteomes.
A. S. haematobium (green spots) vs. S. bovis (red spots).
B. S. haematobium (red spots) vs. E. caproni (green spots).
C. S. bovis (red spots) vs. E. caproni (green spots).
Identified proteins are indicated by solid arrows (for S. haematobium), dashed arrows (for E. caproni) and encircled (for both).

Fig. 2. Pair-wise comparison of protein expression patterns in adult worm proteomes of pairs of parasite species from DIA plug-in analysis. This analysis co-detects the spots from the image and, after normalization, compares the volume of a spot from the two samples as a volume ratio. Spots were detected as different if the volume ratio threshold difference was ⩾ 5 fold. Each gel image shows different expression patterns on individual species gels.
A. Gels showing the DIA analyses of S. haematobium vs. S. bovis on the gels from each of the two species. Green marks represent proteins over-expressed in S. haematobium. Red marks represent proteins over-expressed in S. bovis. Blue represents spots expressed to similar levels in the two species.
B. Gels showing the DIA analyses of S. haematobium vs. E. caproni on the gels from each of the two species. Green marks represent proteins over-expressed in S. haematobium. Red marks represent proteins over-expressed in E. caproni. Blue represents spots expressed to similar levels in the two species.
C. Gels showing the DIA analyses of S. bovis vs. E. caproni on the gels from each of the two species. Green marks represent proteins over-expressed in E. caproni. Red marks represent proteins over-expressed in S. bovis. Blue represents spots expressed to similar levels in the two species.
The histograms represents spot frequencies plotted against logarithm volume ratio.
On the final gel, comparing S. bovis vs. E. caproni, 1757 spots were detected with 78·6% showing similar expression levels. 9·1% of the protein spots showed increased expression in S. bovis and 12·3% showed increased expression in E. caproni (Fig. 2C). It was possible to identify some of the proteins present on the adult worm proteomes by comparing DIGE images with the proteomic maps of S. haematobium and E. caproni. Details of S. haematobium protein identities from mass spectrometry of proteins have already been published (Mutapi et al. Reference Mutapi, Burchmore, Mduluza, Foucher, Harcus, Nicoll, Midzi, Turner and Maizels2005) while those from E. caproni have not previously been published and are given in Table 1 and annotated in Fig. 3.
Table 1. Proteins identified in the adult soluble worm preparation of Echinostoma caproni using MASCOT search engine (Matrix Science)


Fig. 3. Coomassie blue-stained 2-dimensional E. caproni gel showing spots excised, and identified by MASCOT. Corresponding protein identities are given in Table 1.
The expression patterns of pairs of proteins spots between different parasite species are compared in Table 2. The heat shock protein HSP70 was more highly expressed in S. haematobium compared to S. bovis or E. caproni. Between the Schistosoma species, there were similar expression levels of metabolic enzymes, signal transduction molecules and detoxification enzymes, but expression levels of these proteins differed between the latter species and E. caproni. Three homologous proteins were identified in S. haematobium and E. caproni; protein disulfide isomerase, enolase and GST, but the gel migration showed that they differed in molecular weights between the two species.
Table 2. Proteins differentially expressed between the different trematodes identified on the 2-dimensional gel in Differential in Gel Electrophoresis (DIGE)

Immune cross-reactivity
Using sera from hamsters infected with S. haematobium and E. caproni, we performed 2D Western-blot analyses. As expected, homologous pairs of sera and antigen showed the highest levels of recognition (Fig. 4). Interestingly, heterologous sera also detected spots in the gels, confirming cross-reactivity among these trematode species. In this context, S. haematobium sera recognised more antigenic spots in the S. bovis proteome than in E. caproni. There was some cross-reactivity, between E. caproni and S. haematobium. Three spots in the E. caproni proteome which were identified as isoforms of GADPH reacted with sera from both E. caproni-infected and S. haematobium-infected hamsters. However, sera from E. caproni-infected hamsters did not react against GADPH in the S. haematobium proteome.

Fig. 4. Antigen recognition profile of sera from trematode-infected hamsters.
A. S. bovis SWAP antigen recognition by sera from E. caproni-infected hamsters.
B. S. bovis SWAP antigen recognition by sera from S. haematobium-infected hamsters.
C. S. haematobium SWAP antigen recognition by sera E. caproni from infected hamsters.
D. S. haematobium SWAP antigen recognition by sera from S. haematobium-infected hamsters.
E. E. caproni SWAP antigen recognition by sera from E. caproni-infected hamsters.
F. E. caproni SWAP antigen recognition by sera from S. haematobium-infected hamsters.
A novel E. caproni antigen GAPDH is encircled.
DISCUSSION
Trematodes are an evolutionarily distinct group of parasites of importance to both human and veterinary medicine in the diseases that they cause. Understanding similarities and differences in their phenotypic molecular biology is important in several areas such as drug target discovery, vaccine design and development of helminth-derived therapeutic agents for immune disorders; also in the context of when infections are acquired during childhood and beyond. Here, a comparative analysis of the proteome of three species of trematode: S. haematobium, S. bovis and E. caproni using 2D-DIGE was conducted. S. haematobium and S. bovis are closely related species and can undergo hybridisation (Huyse et al. Reference Huyse, Webster, Geldof, Stothard, Diaw, Polman and Rollinson2009), the results obtained here show that there are also significant proteomic differences, even among conserved proteins. These species-specific differences could be due to phenotypic plasticity arising from different host-parasite relationships (Schrimpf and Hengartner, Reference Schrimpf and Hengartner2010). Evolutionary and phylogenetic studies have demonstrated that highly expressed genes tend to evolve more slowly (Hirsh and Fraser, Reference Hirsh and Fraser2001; Schrimpf and Hengartner, Reference Schrimpf and Hengartner2010), nonetheless here we demonstrate that some of these conserved proteins differ in molecular weight, most likely due to post-translational modifications which should be explored further particularly as vaccine candidates.
It was possible to identify some of the proteins present in the proteomes by searching public databases, but due to the limited sequence information available on these three trematodes, a large number of the proteins remain unidentified (Nowak and Loker, Reference Nowak and Loker2005). Comparing the proteomes of the two schistosome species, only HSP70 identified from the Coomassie Blue-stained reference gel showed increased expression in S. haematobium despite the 10% difference in protein expression levels detected by the DIA analysis. The DIGE analysis can detect much lower concentrations of proteins than Coomassie staining. Thus, a large number of proteins present on the DIGE gel were present at a concentration too low to be detected from the Coomassie Blue-stained gel for mass spectrometry. These proteins accounted for some of the 10% differences between the two schistosomes. Our results showing differential expression of HSP70 are consistent with those from a different trematode genus, Fasciola where F. hepatica and F. gigantica show different levels of HSP70 expression (Smith et al. Reference Smith, Spithill, Pike, Meeusen and Piedrafita2008). Furthermore we have previously reported differences in HSP70 expression in E. caproni parasites from low vs. high compatible hosts (Higón et al. Reference Higón, Monteagudo, Fried, Esteban, Toledo and Marcilla2008). Therefore, expression levels of HSP70 seem to depend on the host environment and this could be a common mechanism used by different parasites in order to adapt to different hosts. The sequencing of the genome and subsequent identification of all proteins present in the proteome of all 3 species compared in this study will greatly strengthen such comparative approaches as they will allow more robust comparison of identified proteins as well as comparisons of the number of isoforms and the relative abundance of each isoform to the compared.
There were more differentially expressed proteins between the two different genera. Most proteins identified in both E. caproni and S. haematobium (with known identities) are homologues (protein disulfide isomerase, enolase and GST). However, these homologues have different molecular weights. It is likely that this difference is due to post-translational modifications rather than changes in the gene sequence, since these proteins are highly conserved (Ramajo-Hernández et al. Reference Ramajo-Hernandez, Oleaga, Ramajo-Martın and Perez-Sanchez2007a,Reference Ramajo-Hernández, Pérez-Sánchez, Ramajo-Martín and Oleagab; Sotillo et al. Reference Sotillo, Valero, Sanchez Del Pino, Fried, Esteban, Marcilla and Toledo2008). Furthermore, these proteins are important for the host-parasite relationship (E/S products, immunogenic properties), so these modifications could be involved in the host-parasite surface interaction. Protein disulfide isomerase (PDI) catalyses the formation (oxidation), breakage (reduction) and rearrangement (isomerisation) of disulfide bonds within proteins, thereby permitting their proper folding in the endoplasmic reticulum and transit through the secretory pathway (Ellgaard and Ruddock, Reference Ellgaard and Ruddock2005). PDI has been identified in the E/S products of adult E. caproni, E. friedi and F. hepatica worms, suggesting that it may be important in host-parasite interactions (Salazar-Calderon et al. Reference Salazar-Calderón, Martín-Alonso, Castro and Parra2003; Bernal et al. Reference Bernal, Carpena, Espert, De la Rubia, Esteban, Toledo and Marcilla2006; Sotillo et al. Reference Sotillo, Valero, Sánchez Del Pino, Fried, Esteban, Marcilla and Toledo2010). Moreover, PDI is immunogenic in human S. haematobium infections (Mutapi et al. Reference Mutapi, Burchmore, Mduluza, Foucher, Harcus, Nicoll, Midzi, Turner and Maizels2005) and experimental F. hepatica (Moxon et al. Reference Moxon, Flynn, Golden, Hamilton, Mulcahy and Brophy2010) and it has been shown to be immunologically protective against the hookworm, Ancylostoma (Epe et al. Reference Epe, Behrens, Strube and Schnieder2007). Differences in PDI molecular weight between S. haematobium and E. caproni could be due to post-translational modifications, akin to the PDI glycosylation reported in Trypansoma brucei where it is related to parasite defence (Rubotham et al. Reference Rubotham, Woods, Garcia-Salcedo, Pays and Nolan2005).
The main function of glutathione S-transferase (GST) is detoxification of oxygen and endogenous free radicals (Torres-Rivera and Landa, Reference Torres-Rivera and Landa2008). It is present in Echinostoma spp. and S. bovis tegument and E/S products (Bernal et al. Reference Bernal, Carpena, Espert, De la Rubia, Esteban, Toledo and Marcilla2006; Perez-Sanchez et al. Reference Pérez-Sánchez, Ramajo-Hernández, Ramajo-Martín and Oleaga2006; Sotillo et al. Reference Sotillo, Valero, Sánchez Del Pino, Fried, Esteban, Marcilla and Toledo2010). It is also the leading schistosome vaccine candidate (Capron et al. Reference Capron, Riveau, Capron and Trottein2005; McManus and Loukas, Reference McManus and Loukas2008). There is a difference in the theoretical and observed molecular weights for GST. Ramajo-Hernandez et al. (Reference Ramajo-Hernandez, Oleaga, Ramajo-Martın and Perez-Sanchez2007a) reported no glycosylation of GST in S. bovis. Enolase is a multifunctional glycolytic enzyme (Pancholi, Reference Pancholi2001), also present in E/S products (Bernal et al. Reference Bernal, Carpena, Espert, De la Rubia, Esteban, Toledo and Marcilla2006; Perez-Sanchez et al. Reference Pérez-Sánchez, Ramajo-Hernández, Ramajo-Martín and Oleaga2006; Sotillo et al. Reference Sotillo, Valero, Sánchez Del Pino, Fried, Esteban, Marcilla and Toledo2010). In S. bovis as well as E. caproni, enolase has been identified as a human plasminogen-binding protein; this protein may be involved in preventing blood clotting during feeding in Schistosoma (Ramajo-Hernández et al. Reference Ramajo-Hernández, Pérez-Sánchez, Ramajo-Martín and Oleaga2007b) or in mucosal erosion in Echinostoma (Marcilla et al. Reference Marcilla, Perez-Garcia, Espert, Bernal, Muñoz-Antoli, Esteban and Toledo2007).
To investigate some of the biological differences arising from differences in the proteomes, the immunogenicity of the adult worm antigens was compared. There was cross-reactivity between the three trematode species, but the intensity and antigen pattern recognition patterns differed. The most immune cross-reactivity occurred between the two schistosomes which is consistent with the DIGE results. We have identified a novel antigen for E. caproni, 3 isoforms of GADPH. E. caproni GAPDH was also recognized by sera from S. haematobium-infected hamsters. Interestingly, S. haematobium GADPH antigen was not recognized by sera from E. caproni-infected hamsters. GAPDH's immunogenicity has been reported from other studies and is one of the World Health Organisation's human schistosome vaccine candidates (Bergquist et al. Reference Bergquist, Al-Sherbiny, Barakat and Olds2002; El Ridi et al. Reference El Ridi, Tallima, Mahana and Dalton2010). Nevertheless it has not previously been reported as an antigen in Echinostoma spp. Toledo et al. (Reference Toledo, Espert, Munoz-Antoli, Fried and Esteban2004) discovered an immunogen of 37 kDa, 6 weeks post Echinostoma infections in rats, but the intensity of this response declined during the infection, suggesting that the protein could be released in the juvenile stages of the parasites. This immunogen is likely to be GAPDH and this present study and that of Toledo et al. (Reference Toledo, Espert, Munoz-Antoli, Fried and Esteban2004) suggest that the kinetics of antigen release and antibody production against GAPDH require further investigation, especially in the future context of screening against human sera from infected people.
Schistosomiasis continues to be a major public health problem in several tropical and sub-tropical countries. There are now several studies (e.g. Garba et al. Reference Garba, Barkier, Djibo, Lamine, Sofo, Gouras, Bosque-Oliva, Webster, Stothard, Utzinger and Fenwick2010), showing that children as young as 1 year old are infected and can harbour levels of infection comparable to those in the adults in their communities and the search for an effective vaccine continues to be a key priority (Secor and Colley, Reference Secor and Colley2005). One promising approach being pursued is to treat people, children in particular, repeatedly with praziquantel to induce immune-mediated resistance to re-infection (Black et al Reference Black, Muok, Mwinzi, Carter, Karanja, Secor and Colley2010a). However, studies using this protocol indicate that the number of PZQ treatments required to reduce re-infection is significantly variable and can take several rounds of PZQ treatment (Black et al. Reference Black, Mwinzi, Muok, Abudho, Fitzsimmons, Dunne, Karanja, Secor and Colley2010b). This suggests that an integrated approach using treatment and a recombinant vaccine as proposed by the World Health Organisation (Berquist, Reference Bergquist2004) might lend predictability and consistency as well as improved efficacy to future schistosome control programmes. Thus the molecular phenotypic differences shown in this study, particularly those which appear to be post-translational, may influence the development and production of recombinant vaccines (e.g. bacterial expression systems may not process the proteins appropriately after translation) and affect the efficacy of future vaccines.
Overall this study has demonstrated that, despite several biological and phylogenetic similarities between the three trematode species S. haematobium, S. bovis and E. caproni, there are quantitative and qualitative differences in protein expression patterns in their adult worm proteomes. The differences could be due to phenotypic plasticity arising from different host-parasite relationships. Some of these differences translate to differences in immunogenicity. Further studies characterizing the differentially expressed proteins will be important in determining the identity of proteins involved in host-parasite adaptation and the nature of the interaction between the host and parasite. This is particularly important for identifying vaccine candidates and predicting the effects vaccination, especially in childhood, would have on the parasite population structure.
ACKNOWLEDGEMENTS
We are grateful for the donation of CyDye DIGE Fluor minimal dyes by GE Healthcare. We also thank Alan Scott, University of Glasgow for technical support.
FINANCIAL SUPPORT
This work was supported by the Carnegie Trust for the Universities of Scotland; the Wellcome Trust (Grant no WT082028MA); and a predoctoral fellowship from the Spanish Ministry of Science and Education (MH).







