Introduction
Sponges (Phylum Porifera) are often an abundant and diverse component of several marine ecosystems (van Soest et al., Reference van Soest, Boury-Esnault, Vacelet, Dohrmann, Erpenbeck, de Voogd, Santodomingo, Vanhoorne, Kelly and Hooper2012). They are filter-feeding, sessile organisms with a simplistic body plan and are considered among the earliest of multicellular animals (Wörheide et al., Reference Wörheide, Dohrmann, Erpenbeck, Larroux, Maldonado, Voigt, Borchiellini and Lavrov2012). The phylum covers a total of 9641 accepted marine species (de Voogd et al., Reference de Voogd, Alvarez, Boury-Esnault, Cárdenas, Díaz, Dohrmann, Downey, Goodwin, Hajdu, Hooper, Kelly, Klautau, Lim, Manconi, Morrow, Pinheiro, Pisera, Ríos, Rützler, Schönberg, Turner, Vacelet, van Soest and Xavier2024) found over a wide range of habitats from the eulittoral zone to hadal depths and from the tropics to the poles (van Soest et al., Reference van Soest, Boury-Esnault, Vacelet, Dohrmann, Erpenbeck, de Voogd, Santodomingo, Vanhoorne, Kelly and Hooper2012). With the exception of carnivorous sponges, all sponges contain specialised cells known as choanocytes or collar cells, which consist of a flagellum surrounded by tentacles. The flagellum creates a pressure gradient drawing in large volumes of water through openings known as ostia. The water containing nutrients, microorganisms, and organic matter (dissolved and particulate) is transported throughout the sponge tissues (Maldonado et al., Reference Maldonado, López-Acosta, Sitjà, García-Puig, Galobart, Ercilla and Leynaert2019).
In addition to consuming microorganisms, sponges also establish symbiotic relationships whereby the symbionts play critical roles in enabling their host sponges to survive and thrive (Wilkinson, Reference Wilkinson1984). To achieve this, the symbionts perform a wide range of functions varying from nutrient translocation to the synthesis of secondary metabolites, which play important roles in host defence and competition with other benthic organisms (Thakur et al., Reference Thakur, Anil and Müller2004; Freeman et al., Reference Freeman, Thacker, Baker and Fogel2013; Thinesh et al., Reference Thinesh, Meenatchi, Lipton, Anandham, Jose, Tang, Seghal Kiran and Selvin2020). They also produce antibiotic compounds, which can inhibit colonisation by potentially pathogenic microorganisms (Piel, Reference Piel2009; Fan et al., Reference Fan, Reynolds, Liu, Stark, Kjelleberg, Webster and Thomas2012). The secondary metabolites and antibiotics are also of biotechnological and/or pharmaceutical interest making sponges one of the most bio-prospected of marine organisms (Blunt and Munro, Reference Blunt and Munro1998; Osinga et al., Reference Osinga, Tramper and Wijffels1998; van Soest and Braekman, Reference van Soest and Braekman1999; Faulkner et al., Reference Faulkner, Harper, Haygood, Salomon, Schmidt and Fusetani2000; Sipkema et al., Reference Sipkema, Franssen, Osinga, Tramper and Wijffels2005; Taylor et al., Reference Taylor, Radax, Steger and Wagner2007). Much of these compounds are produced by the microbial symbionts, which has helped to drive the interest in studying sponge-associated microbial communities (Taylor et al., Reference Taylor, Radax, Steger and Wagner2007; Cleary et al., Reference Cleary, Becking, Pires, de Voogd, Egas and Gomes2013, Reference Cleary, de Voogd, Polónia, Freitas and Gomes2015, Reference Cleary, Swierts, Coelho, Polónia, Huang, Ferreira, Putchakarn, Carvalheiro, van der Ent, Ueng, Gomes and de Voogd2019; Thomas et al., Reference Thomas, Moitinho-Silva, Lurgi, Björk, Easson, Astudillo-García, Olson, Erwin, López-Legentil, Luter, Chaves-Fonnegra, Costa, Schupp, Steindler, Erpenbeck, Gilbert, Knight, Ackermann, Victor Lopez, Taylor, Thacker, Montoya, Hentschel and Webster2016; Swierts et al., Reference Swierts, Cleary and de Voogd2018; Cleary et al., Reference Cleary, Polónia and de Voogd2018a).
The present study is a preliminary investigation of the sponge fauna, sponge-associated, and planktonic prokaryotic communities residing in Burgers' Zoo Ocean aquarium, which is located in Arnhem, the Netherlands. The aquarium system contains eight million litres of artificial seawater dispersed among 10 large tanks, which mimic various aspects of the flora and fauna of the Indo-Pacific Ocean. Burgers' Zoo Ocean aquarium opened to the public in the year 2000 and one of these tanks has maintained a tropical living coral reef ecosystem (Janse et al., Reference Janse, Wensing, Gieling, De Jongh, Leewis and Janse2008). This tank has a total volume of 750,000 L and contains varying reef substrates including rocks, sand, corals and coral rubble and has a maximum depth of 6 m. Our main goals in the present study were to (1) identify the sponges growing in the aquarium, (2) compare prokaryotic diversity and composition among sponge species and water, (3) identify the most abundant operational taxonomic units (OTUs) and closely related organisms using BLAST, (4) compare predicted gene counts for selected KEGG (Kyoto Encyclopaedia of Genes and Genomes) categories among sponge species and water samples, and (5) identify potential pathogens and beneficial microorganisms inhabiting the aquarium system.
Material and methods
Burgers' Zoo Aquarium
The ecological and physicochemical features of the aquarium system have been described in detail by Janse et al. (Reference Janse, Wensing, Gieling, De Jongh, Leewis and Janse2008) and Ernst et al. (Reference Ernst, Janse, Renema, Kouwenhoven, Goudeau and Reichart2011). In short, the aquarium system is maintained using artificial seawater (Tropic Marin® Zoomix, Hünenberg, Switzerland), which is prepared with a fixed salinity of 34 ppt. Salinity, temperature, and pH are monitored frequently and range between 33.9 and 34.3 ppt, 24.5 to 26.5°C, and 7.90–8.10, respectively (Janse et al., Reference Janse, Wensing, Gieling, De Jongh, Leewis and Janse2008; Ernst et al., Reference Ernst, Janse, Renema, Kouwenhoven, Goudeau and Reichart2011). Certain changes have been made to the aquarium system following the publication of Janse et al. (Reference Janse, Wensing, Gieling, De Jongh, Leewis and Janse2008). For example, the high-rate sand filters have been replaced by a drum filter (Profidrum® 75/80 with 40 μm screen). Water quality management has also intensified over the years, particularly as concerns the addition of macro-elements such as calcium (range 400–440 mg Ca2+/L), magnesium (range 1300–1350 mg Mg2+/L), and strontium (range 8–10 mg Sr2+/L). Alkalinity has been kept between 2.7 and 3.2 mEq/L. Nutrients in the aquarium system are managed by the amount of food fed to the system, the number of fish, and by addition of a carbon source (diluted ethanol at 30 g C/d) in order to increase the growth of heterotrophic bacteria within the water column. The increase in bacterial biomass has fixed the dissolved N and P in the water and removed the ortho-phosphates from the water; ortho-phosphate levels were kept between 0.01 and 0.06 mg PO43−/L, and nitrate levels between 0.4 and 2.2 mg NO3−/L.
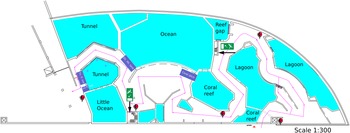
The aquarium (Figure 1) is considered an Indo-Pacific coral reef aquarium as it only contains biota native to the Indo-Pacific region. Presently the aquarium houses approximately 112 scleractinian corals, 10 octocorals, and 68 fish species. The specific origin of the reef fauna varies as most of the corals come from other public aquaria or were confiscated by the Dutch Government from illegal imports from the Indo-Pacific, whereas the live rock and associated benthos were mostly collected from shallow-water reefs surrounding the islands of Java and Bali, Indonesia (Ernst et al., Reference Ernst, Janse, Renema, Kouwenhoven, Goudeau and Reichart2011). It is likely that the sponges, which colonised the aquarium system, originated from live rock segments. The tanks have their own filtration systems, which run on a continuous basis. The ‘fish only’ tanks get 10–25% of their volume in new water per year. This water originates from the live coral reef tank, which should be the base of dispersal of sponges over the different tanks. The live coral reef tank receives 120–140% of its volume in new water each year.

Figure 1. Schematic drawing of Burgers' Zoo Ocean aquarium. The tanks coloured purple are not drawn to scale and represent tanks on a different floor level to the public display tanks. These tanks are: tunnel pre-filtration tank (Tn. fl. tnk), little ocean pre-filtration tank (LO fl. tnk), ocean filtration tank (Oc. flt. Tnk.) and coral ditch.
Filtration for the tunnel and little ocean tanks consists of pre-filtration tanks, which are filled by gravity from the display tanks. From the pre-filtration tanks, water is brought to the second floor with a pump. 50% of the tunnel flow goes through a drum filter (Profidrum®, Lopik, Netherlands type 65 with 40 μm screen at 70 m3/h), followed by two parallel protein skimmers (Schuran®, Jullich, Germany, Aquafloater AQ 800, 25 m3/h each), and two custom made trickling filters; the other 50% goes directly through two parallel protein skimmers (Aquafloater AQ 800) and two custom made trickling filters. The little ocean tank (Figure 1) consists of a total flow of 70 m3/h, with a drum filter (Profidrum, type 55), two parallel protein skimmers (Aquafloater AQ 800, 25 m3/h each), and two custom made trickling filters. The lagoon tank consists of two parallel protein skimmers (Aquafloater AQ 800, 25m3/h each), two custom made trickling filters, and a Triton TR 100 high rate sand filter (Pentair®, 10 m3/h) with AFM filter media (Dryden Aquaculture, Bonnyrigg, United Kingdom). Just after finishing this study, a UV-C system (VGE®, Schijndel, Netherlands at 40 m3/h) was added to the lagoon tank. The reef gap tank filtration consists of a drum filter (Profidrum type 45 with 40 μm screen at 20 m3/h), followed by protein skimmers (Schuran AQ 500 at 15 m3/h), and a custom made trickling filter.
The different tanks vary in their respective water filtering methods and several measured biogeochemical parameters (nitrate, phosphate, total organic carbon, TOC, and dissolved organic carbon, DOC concentrations; Supplementary data 1). The little ocean and tunnel tanks have a separate phosphate filter where phosphates were removed by chemical precipitation by adding FeCl3 solution. The lagoon, reef gap, and little ocean tanks were treated with a denitrification filter with autotrophic Sulphur bacteria (including Thiobacillus thiooxidans) and elemental sulphur. The pre-filtration tank of the little ocean receives water from the aquarium and the effluent of both the phosphate filter and sulphur filter. The tunnel is treated with a denitrification filter with heterotrophic, anaerobic bacteria (multiple species) and methanol (CH3OH). The pre-filtration tank of the tunnel receives water from the aquarium and the effluent of both the phosphate filter and denitrification filter.
The coral reef ditch is a 6.0 × 1.0 × 0.3 m tank and is part of the living coral reef tank. The coral reef tank has little external filtration consisting of a drum filter (Profidrum type 65 with 40 μm at 70 m3/h) and two parallel protein skimmers (Schuran ® type AQ 800 and AQ 500). Most biological activity happens within the living ecosystem (Janse et al., Reference Janse, Wensing, Gieling, De Jongh, Leewis and Janse2008). The coral reef system is fed with ethanol (C2H5OH) within the water column to increase heterotrophic bacterial build-up within the water. Nitrate and phosphate were measured using a HACH spectrophotometer (DR 3900; HACH®, Ames, Iowa, USA) via methods 8192 and 8039 for nitrate and 8048 for phosphate. TOC and DOC were measured by Triton laboratory (Fernwald, Germany). Results of biochemical parameters are found in Supplementary data 1.
Sampling
A total of 19 samples consisting of seven sponge species and five water samples were collected from the aquarium system (Supplementary data 2); a schematic design of the system is presented in Figure 1 and pictures of tanks and sponges in Figure 2. The tanks are identified as the lagoon, reef gap, tunnel (both tunnel aquarium and tunnel pre-filtration tank), little ocean pre-filtration tank, and coral reef ditch, a tank adjacent to the coral reef aquarium.

Figure 2. Images taken at Royal Burgers' Zoo Ocean aquarium. (A) Coral Reef display, (B) Ocean filtration tank, (C) Chondrosia aff. corticata, (D) Suberites aff. diversicolor, (E) Chondrilla aff. australiensis, (F) Cinachyrella sp. specimen in the tunnel, (G) Stylissa carteri, and (H) Chalinula aff. milnei.
Sponge samples were identified by NJdV using classical morphological methods and voucher specimen have been incorporated into the sponge collection of Naturalis Biodiversity Center. Water samples were collected from the lagoon, reef gap, tunnel, little ocean pre-filtration tanks, and coral reef ditch by filtering 1 L of water through a Millipore White Isopore Membrane Filter (GTTP04700, 47 mm diameter, 0.22 μm pore size; Merck Millipore, Sigma, Burlington, MS, USA). Sponge samples and membrane filters were stored in 96% EtOH for subsequent analysis.
DNA extraction and next-generation sequencing analysis
PCR-ready genomic DNA was isolated from all samples using the FastDNA® SPIN soil Kit (MPbiomedicals) following manufacturer's instructions. Briefly, the whole membranes used for water samples were cut into small pieces and transferred to Lysing Matrix E tubes containing a mixture of ceramic and silica particles; for the sponges ± 500 mg of tissue (including parts of the surface and interior) was added. The microbial cell lysis was performed in the FastPrep® Instrument (Q Biogene) for 80 s at 6.0 ms−1. The extracted DNA was eluted into DNase/pyrogen-free water to a final volume of 50 μl and stored at −20°C until use. The 16S rRNA gene V3-V4 variable region PCR primers 341F 5′-CCTACGGGNGGCWGCAG-3′ and 785R 5′-GACTACHVGGGTATCTAATCC-3′ (Klindworth et al., Reference Klindworth, Pruesse, Schweer, Peplies, Quast, Horn and Glöckner2013) with barcode on the forward primer were used in a 30 cycle PCR assay using the HotStarTaq Plus Master Mix Kit (Qiagen, USA) under the following conditions: 94°C for 3 min, followed by 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, after which a final elongation step at 72°C for 5 min was performed. After amplification, PCR products were checked on a 2% agarose gel to determine the success of amplification and the relative intensity of bands; the blank control did not yield any bands. PCR product was used to prepare the DNA library following the Illumina TruSeq DNA library preparation protocol. Next-generation, paired-end sequencing was performed at MrDNA (Molecular Research LP; http://www.mrdnalab.com/; last checked 18 November 2016) on an Illumina MiSeq device (Illumina Inc., San Diego, CA, USA) following the manufacturer's guidelines. Sequences from each end were joined following Q25 quality trimming of the ends followed by reorienting any 3′–5′ reads back into 5′–3′ and removal of short reads (<150 bp).
16S rRNA Gene Sequencing Analysis
The 16S rRNA gene amplicon libraries were analysed using QIIME2 (version 2019.7; Bolyen et al., Reference Bolyen, Rideout, Dillon, Bokulich, Abnet, Al-Ghalith, Alexander, Alm, Arumugam, Asnicar, Bai, Bisanz, Bittinger, Brejnrod, Brislawn, Brown, Callahan, Caraballo-Rodríguez, Chase, Cope, Da Silva, Diener, Dorrestein, Douglas, Durall, Duvallet, Edwardson, Ernst, Estaki, Fouquier, Gauglitz, Gibbons, Gibson, Gonzalez, Gorlick, Guo, Hillmann, Holmes, Holste, Huttenhower, Huttley, Janssen, Jarmusch, Jiang, Kaehler, Kang, Keefe, Keim, Kelley, Knights, Koester, Kosciolek, Kreps, Langille, Lee, Ley, Liu, Loftfield, Lozupone, Maher, Marotz, Martin, McDonald, McIver, Melnik, Metcalf, Morgan, Morton, Naimey, Navas-Molina, Nothias, Orchanian, Pearson, Peoples, Petras, Preuss, Pruesse, Rasmussen, Rivers, Robeson, Rosenthal, Segata, Shaffer, Shiffer, Sinha, Song, Spear, Swafford, Thompson, Torres, Trinh, Tripathi, Turnbaugh, Ul-Hasan, van der Hooft, Vargas, Vázquez-Baeza, Vogtmann, von Hippel, Walters, Wan, Wang, Warren, Weber, Williamson, Willis, Xu, Zaneveld, Zhang, Zhu, Knight and Caporaso2019). Raw data were imported yielding a demultiplexed ‘qza’ data file (artefact). The DADA2 plugin (Callahan et al., Reference Callahan, McMurdie, Rosen, Han, Johnson and Holmes2016) in QIIME 2 was subsequently used to trim sequences (final length 400 nt). The DADA2 analysis yielded output archives containing an operational taxonomic unit (OTU, also known as amplicon sequence variant or ‘ASV’) table, denoising stats, and a fasta file of representative sequences. The feature-classifier plugin with the extract-reads method was then used with the i-sequences argument set to silva-138-99-seqs.qza. This was followed by the feature-classifier plugin with the fit-classifier-naive-bayes method and the i-reference-taxonomy method set to silva-138-99-tax.qza. Both silva-138 files can be obtained from https://docs.qiime2.org/2020.8/data-resources/?highlight=silva. The feature-classifier plugin was then used with the classify-sklearn method and the i-reads argument set to the representative sequences file generated by the DADA2 analysis to produce a table with taxonomic assignments for all OTUs. Finally, mitochondria, chloroplasts, and Eukaryota were filtered out using the QIIME 2 taxa plugin with the filter-table method. The OTU and taxonomy tables were later merged in R. After quality control, rarefaction, and removal of OTUs assigned to chloroplasts and mitochondria, the dataset consisted of 2,005,389 sequences and 8471 OTUs. An OTU counts table is presented in Supplementary data 3. The 50 most abundant OTUs are presented in Supplementary data 4. Accession numbers of closely related organisms in GenBank to selected OTUs were obtained using NCBI Basic Local Alignment Search Tool (BLAST) (Altschul et al., Reference Altschul, Gish, Miller, Myers and Lipman1990).
Predicted metagenomic analysis
In the present study, the Tax4Fun2 library (Wemheuer et al., Reference Wemheuer, Taylor, Daniel, Johnston, Meinicke, Thomas and Wemheuer2020) in R was used to predict the metagenomic content of each sample with the KEGG (Kyoto Encyclopaedia of Genes and Genomes) database. First, the runRefBlast function with the database mode was set to ‘Ref100NR’ and the path_to_otus argument set to the representative sequences file generated using QIIME2. The makeFunctionalPrediction argument was then used with the path_to_otu_table argument set to the OTU table and the min_identity_to_reference argument set to 0.95. This entails predicting function for a given OTU at approximately generic level. A number of studies have shown that traits tend to be conserved at or below family level (Owens and Bennett, Reference Owens and Bennett1995; Peterson et al., Reference Peterson, Soberóna and Sánchez-Cordero1999). Prokaryotic families also have distinct metabolic capabilities that are also prevalent at the family level and below (Goldford et al., Reference Goldford, Lu, Bajić, Estrela, Tikhonov, Sanchez-Gorostiaga, Segrè, Mehta and Sanchez2018). Default settings were used for the other arguments. Output of Tax4Fun2 consists of tables of functional counts for individual pathways and KEGG orthologs (KOs). Because of functional overlap, some KEGG orthologs (KOs) can be represented in multiple pathways. Note that the Tax4Fun2 results as presented are predictive and thus provide information on potential enrichment and putative function as opposed to measuring actual gene presence/expression and function.
Statistical analyses
A table containing the OTU counts was imported into R. Supplementary data 3 contains all operational taxonomic unit (OTU) counts per sample, including the OTU number and ASV codes, in addition to taxonomic classifications for all OTUs. The raw OTU counts matrix was used to compare diversity among sponge species and water samples and to investigate differences in higher taxon abundance. Diversity indices were obtained using the rarefy and diversity functions from the vegan (Oksanen et al., Reference Oksanen, Blanchet, Friendly, Kindt, Legendre, McGlinn, Minchin, O'Hara, Simpson, Solymos, Stevens, Szoecs and Wagner2020) package in R (R Core Team, 2022). Variation in prokaryotic composition among biotopes (sponge species and water samples from different compartments) was visualised with Principal Coordinates Analysis (PCO). For the PCO, a Bray-Curtis distance matrix was first obtained using the Phyloseq package (McMurdie and Holmes, Reference McMurdie and Holmes2013) whereby the count data was rarefied using the rarefy_even_depth function with the sample size argument set to the minimum sample size (38,608 in the present study) and subsequently log10 transformed. Finally, we tested for a significant difference in prokaryotic composition between sponge and water samples with an adonis analysis from the vegan package.
Results and discussion
Overview of sponge fauna in the Ocean aquarium
In the present study, seven sponge species, belonging to seven different genera and six orders, were sampled from Burgers' Zoo Ocean aquarium. Chondrosia aff. corticata Thiele, 1900 (Order Chondrosida, Family Chondrillidae) (Figure 2C) grew as thin spreading mats across the pipes in the outlet of the pre-filtration tanks of the tunnel and little ocean tank. The samples were very slippery and cream coloured. Sponges belonging to the genus Chondrosia are extremely difficult to identify because of the lack of an inorganic skeleton. Our specimens had thick cortices, dense choanosomes without any foreign inclusions, and only some small, dark pigmented, spherulous cells. Only three species are known from the Indo-Pacific region, namely, Chondrosia chucalla, C. corticata, and C. tenochca. Chondrosia tenochca is a dark-coloured, encrusting species and also has a dense skeleton without foreign elements. Chondrosia corticata, originally described from the Moluccas, Indonesia, was recently recorded from Mozambique and had, unlike our specimens, a cavernous choanosome, large protruding oscules, and a brown colour. We cannot determine with certainty the true identity of the specimens, but they resembled this species.
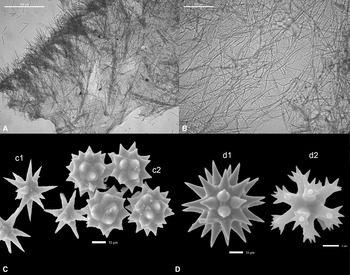
Both Chondrilla aff. australiensis Schulze, 1877 (Order Chondrosida, Family Chondrillidae) samples (Figure 2E) were white, formed firm lobes with numerous small oscules, and grew in the pre-filtration tanks of the tunnel and little ocean. The ectosome was characterised by a dense ectosomal layer of spicules aggregated around the canals. The spicules were composed of both oxysphaerasters (Figure 3C) and oxyasters in similar dimensions. The oxysphaerasters had short, thick mamillate rays varying in size from 18 to 26 μm with a mean of 23 μm. The oxyasters (varying in size from 20 to 28 μm with a mean of 22 μm) had tapering slender rays, which had a roughened surface. Only three Chondrilla species are known from the Indo-Pacific region, which have oxysphaerasters and smaller oxyasters as a spicule complement, and have similar distributions from the Red Sea and Indian Ocean to the western Pacific Ocean. The identification of Chondrilla species is also extremely difficult due to the lack of discriminating characters, their large distributional range, and possible presence of sympatric species. Chondrilla australiensis Carter 1873 is a brown encrusting sponge, originally described from Australia. The oxysphaerasters (varying in size from 20 to 30 μm with a mean of 25 μm) had short thick rays abruptly tapering to points and these were occasionally mammilate. The small oxyasters (varying in size from 20 to 25 μm with a mean of 22 μm) had tapering microspined rays, which were frequently bifurcate or multilayered.

Figure 3. A. Suberites aff. diversicolor cross section, (B) Chalinula aff. milnei cross section, (C) Chondrilla aff. australiensis, SEM image of spicules, oxyaster (c1), oxysphaeraster (c2), (D) Tethya aff. microstella, SEM image of microscleres, megasters (d1), and micrasters (d2).
Chondrilla mixta Schulze, 1877 was originally described from the Red Sea and the type has not been examined by any of the reports available. The validity of this species is, therefore, questionable, but the spherasters are supposed to be smaller than those of C. australiensis. This species was recently reported from Hawaii (Núñez Pons et al., Reference Núñez Pons, Calcinai and Gates2017). The morphology and size of the sphaerasters (12–50 μm) and oxysphaerasters (12–20 μm) differs from our species. Chondrilla jinesis Hentschel, 1912 is an encrusting, grey to black species described from Indonesia. The oxysphaerasters are 48–56 μm and have very thin, large oxyasters (45–80 μm). This species has not been reported since its description. Although our specimens had different colours, overall habitus, and spicule morphologies than Chondrilla australiensis, they were most similar to this species.
We collected two specimens of Suberites aff. diversicolor Becking & Lim, 2009 (Order Suberitida, Family Suberitidae) from the reef gap tank (Figure 2D). The species was light-yellow and thickly encrusting to massive. The skeleton consisted of smaller tylostyles arranged in bouquets in the outer part of the sponges and there was some radial orientation visible, which became more confusing in the interior of the sponges (Figure 3A). Most of the heads of the tylostyles had modifications and the size of the spicules was approximately 175 μm up to 880 μm in length. Presently, more than 80 species of Suberites have been described worldwide, but only seven from the Indo-Pacific region. The spicule dimensions of the specimen from Burgers' Zoo aquarium were very similar to those of Suberites diversicolor, which has been described from anchialine lakes and mangrove roots. Suberites diversicolor is variable in habit and colour and also has a wide distributional range including Indonesia, Vietnam, Singapore, and Australia.
Only a single specimen of Tethya aff. microstella Sarà, 1990 (Order Tethyida, Family Tethyidae) was observed in the pre-filtration tank of the little ocean, which was white. The spicules were composed of strongyloxeas (varying in size from 662 to 1985 μm with a mean of 1402 μm), megasters with a thick centrum (varying in size from 48 to 73 μm with a mean of 60 μm), and very small micrasters (6–7 μm) (Figure 3D). Only a few Tethya species are known to have such small micrasters. Interestingly, Tethya sponges are often observed in aquaria and several new species have been described from aquaria. Sarà et al. (Reference Sarà, Sarà, Nickel and Brümmer2001) described three new species from a public German aquarium of presumed Indo-Pacific origin. These species were also white, which is unusual for Tethya species. The morphology and size of the spherasters and strongylasters of T. minuta were very similar to our species, however the strongyloxeas (800–1350 μm) were smaller than in T. minuta and our specimen lacked the nuclear styles. The brownish to yellowish Tethya microstella, described from Orpheus island, Great Barrier Reef, is another Tethya species with small micrasters. It is characterised by having very small tylasters (4–6 μm) that only occur in the ectosome as in our species and also the megasters have a similar morphology and dimensions (30–80 μm). The strongyloxeas (400–1600 μm) are slightly shorter than those of our specimen. Because of the similarity of the spicule composition, our specimen is most similar to Tethya microstella, but deviates in the dimensions of the spicules. This species was reported from the Maldives as well, although the dimensions of the spicules were even smaller than those of the original species.
Several very small, possibly juvenile, specimens of golf ball (Cinachyrella sp.; Order Tetractinellida, Family Tetillidae) sponges were collected from the lagoon tank (Figure 2F). The identification of these specimens is challenging because of their small size. In general golf ball sponges tend to be highly adaptable to varying environmental parameters and can be found across a wide range of habitats all around the globe (de Voogd and Cleary, Reference de Voogd and Cleary2008; de Voogd et al., Reference de Voogd, Becking and Cleary2009; Cleary et al., Reference Cleary, Becking, Pires, de Voogd, Egas and Gomes2013; Cleary et al., Reference Cleary, Polónia and de Voogd2018a). In a recent publication, Cleary and de Voogd (Reference Cleary and de Voogd2024) used 28s Barcoding for several species of golf ball sponges, including the specimens from Burgers' Ocean aquarium. Our specimens were characterised by having very small sigmaspires varying in size from 4.6 to 9.8 μm with a mean of 7 μm, oxeas varying from 800 to 2262 μm, and very long and thin ana- and protrianes, which were mostly broken. The longest measured over 3000 μm in length. The specimens lacked acanthose microxeas, which is a notable character of the widespread Indo-Pacific sponge Cinachyrella australiensis. Cinachyrella albatridens Lendenfeld 1907 described from the deep sea near Chagos in the Indian Ocean and C. arabica from Oman also have very small sigmaspires (7–12 μm) and are probably closely related to our species. However, our oxeas were much smaller, and the trianes were longer. Our species did not fit any of the existing species described thus far.
A single specimen of the common and widespread Indo-Pacific sponge species Stylissa carteri (Dendy, 1889) (Order Scopalinida, Family Scopalinidae) was collected from the coral reef ditch tank. The specimen was confiscated from an illegal collection at Amsterdam airport and transported to Burgers' Zoo. The species was bright orange and had a flabellate growth form. The spicules were thick styles and varied from 484 to 615 μm in length and 18 to 25 μm in width. This species was originally described from the Gulf of Mannar and can be found throughout the Indo-Pacific region, but it has recently been suggested that Stylissa carteri constitutes a cryptic species complex (Erpenbeck et al., Reference Erpenbeck, Aryasari, Benning, Debitus, Kaltenbacher, Al-Aidaroos, Schupp, Hall, Hooper, Voigt, de Voogd and Wörheide2017).
We collected three specimens of Chalinula aff. milnei (De Laubenfels, 1954) (Order Haplosclerida, Family Chalinidae) (Figure 2H) from the tunnel. This sponge grew profusely across artificially constructed corals and walls of the lagoon and tunnel but not in other areas of the aquarium system. The specimens were thinly encrusting and grey to bluish. The specimens were very similar to Chalinula milnei originally described from the Ebon Atoll (Marshall islands). Chalinula milnei has thin strongyloxeas with irregular endings with a size range of 73 to 114 μm in length and 1.2 to 3.3 μm in width. The skeleton is typical for the genus Chalinula with secondary lines in the choanosome (Figure 3B) of more than one spicule length and the absence of an ectosomal skeleton. Chalinula milnei is known to actively overgrow and smother live corals (Hoeksema et al., Reference Hoeksema, Dekker and de Voogd2014). Although the lagoon in the aquarium only contained artificially constructed coral, the sponge can pose a potential risk of entering the live coral reef.
Prokaryotic communities identified in aquarium sponges and water
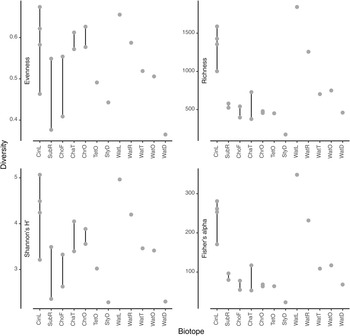
The most abundant prokaryotic phyla (in terms of sequences) in the present study were Proteobacteria (957,371 sequences; 47.7% of all sequences; 3196 OTUs), Bacteroidota (281,595 sequences; 14.0% of all sequences; 1119 OTUs), Campilobacterota (154,215 sequences; 7.7% of all sequences; 75 OTUs), and Actinobacteriota (148,978 sequences; 7.4% of all sequences; 253 OTUs). Boxplots of evenness (Pielou's J), rarefied richness, Shannon's H' and Fisher's alpha diversity indices are shown in Figure 4. Among water samples, OTU richness and evenness were highest in water from the lagoon (WatL) and reef gap (WatR) tanks, intermediate in the tunnel (WatT) and little ocean pre-filtration tank (WatO), and lowest in the coral reef ditch (WatD).

Figure 4. Values for selected diversity indices (a) Evenness, (b) Richness, (c) Shannon's H’ and (d) Fisher's alpha. The biotopes sampled were: CinL, Cinachyrella sp.; SubR, Suberites aff. diversicolor; ChoF, Chondrosia aff. corticata; ChaT, Chalinula aff. milnei; ChrO, Chondrilla aff. australiensis; StyD, Stylissa carteri.; TetO, Tethya aff. microstella; WatL, water lagoon; WatR, water reef gap; WatT, water tunnel; WatO, Little ocean pre-filtration tank; WatD, water coral reef ditch. Note that for the sponges, the final letter of the code indicates the tank in which they were sampled, namely, L for lagoon, R for reef gap, T for tunnel, O for ocean pre-filtration tank, and D for coral reef ditch.
Among sponge species, OTU richness was highest in Cinachyrella sp. and lowest in Stylissa carteri reflecting the pattern in water. Evenness was variable, but highest in the sponge species Cinachyrella sp., Chalinula aff. milnei, and Chondrilla aff. australiensis (Figure 4). Figure 5 shows the mean relative abundances of the eight most abundant phyla in each biotope accounting for 90.18% of all sequences and 62.93% of all OTUs. Of note are the markedly higher levels of Proteobacteria in water from the lagoon and reef gap tanks, higher levels of Bacteroidota in water from the tunnel and little ocean pre-filtration tank, and high levels of Campilobacterota in water from the coral reef ditch. Campilobacterota abundance was also relatively high in S. carteri and Chalinula aff. milnei. Acidobacteriota and Gemmatimonadota were most abundant in the sponge species Chondrosia aff. corticata and Chondrilla aff. australiensis whereas Actinobacteriota abundance was greatest in the sponge species Chondrilla aff. australiensis and Tethya aff. microstella and also relatively high in Cinachyrella sp.. Chloroflexi abundance was relatively low in all biotopes reaching greatest abundance in Chondrilla aff. australiensis at just over 10%. NB1-j was rare in all biotopes except Cinachyrella sp..

Figure 5. Barplots of the mean relative abundances of the eight most abundant phyla for the following biotopes: CinL, Cinachyrella sp.; SubR, Suberites aff. diversicolor; ChoF, Chondrosia aff. corticata; ChaT, Chalinula aff. milnei; ChrO, Chondrilla aff. australiensis; StyD, Stylissa carteri.; TetO, Tethya aff. microstella; WatL, water lagoon; WatR, water reef gap; WatT, water tunnel; WatO, Little ocean pre-filtration tank; WatD, water coral reef ditch. The bars are colour-coded according to the phylum as indicated by the legend in the topright corner. Note that for the sponges, the final letter of the code indicates the tank in which they were sampled, namely, L for lagoon, R for reef gap, T for tunnel, O for ocean pre-filtration tank, and D for coral reef ditch.
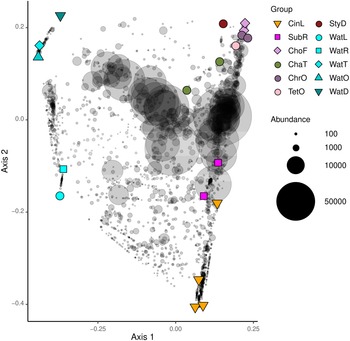
A PCO ordination is presented in Figure 6. Eigenvalues for the first two axes of variation were 1.12 for axis 1 and 0.89 for axis 2, which together explained 34% of the variation in composition. There was a highly significant difference in composition between sponge and water samples (P 1,17 = 3.91, P < 0.001, R 2 = 0.813). In line with this, the main axis of variation separated sponge from water samples whereas the second axis separated sponge species collected in the lagoon and reef gap tanks from sponge species collected in the remaining tanks. The water in which the sponges resided, thus, appeared to have an effect on prokaryotic composition. The species Chondrilla aff. australiensis and Chondrosia aff. corticata sampled in the tunnel and little ocean pre-filtration tanks also clustered together, and clustered closer to other sponge specimens sampled in these tanks. Specimens of Cinachyrella sp. and Suberites aff. diversicolor sampled from the lagoon and reef gap tanks also clustered together along the second PCO axis. The lagoon and reef gap tanks did not house any live corals, but different fish species, and the illumination was lower.

Figure 6. Ordination showing the first two axes of the principal coordinates analysis (PCO) of OTU composition. Symbols are colour-coded and represent samples from different biotopes as shown in the legend on the right side of the figure. Grey symbols represent weighted averages scores for OTUs. The symbol size is proportional to biotope abundance (number of sequence reads). The legend symbols represent the following biotopes: CinL, Cinachyrella sp.; SubR, Suberites aff. diversicolor; ChoF, Chondrosia aff. corticata; ChaT, Chalinula aff. milnei; ChrO, Chondrilla aff. australiensis; StyD, Stylissa carteri; TetO, Tethya aff. microstella; WatL, water lagoon; WatR, water reef gap; WatT, water tunnel; WatO, Little ocean pre-filtration tank; WatD, water coral reef ditch. Note that for the sponges, the final letter of the code indicates the tank in which they were sampled, namely, L for lagoon, R for reef gap, T for tunnel, O for ocean pre-filtration tank, and D for coral reef ditch.
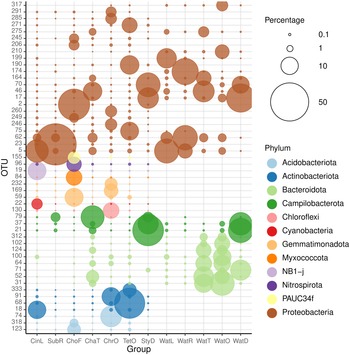
The 50 most abundant OTUs (Figure 7) can be divided into several categories based on their distributions among the biotopes studied and the results of BLAST analyses (Supplementary data 4). A large number of OTUs were mainly found in water samples. These included OTUs 31, 37, 62, 64, 71, 100, 102, 124, 164, 174, 199, 210, 312, and 317. All of these OTUs were related to microorganisms in the GenBank database mainly obtained from seawater, but also pond water, biofilm in a cold seep, oil, and the algal species Sargassum vachellianum. All of these OTUs had ≥99% sequence similarities to microorganisms in GenBank with the exception of OTU-199, which had only 94.7% sequence similarity (Supplementary data 4). OTUs 31, 64, 71, 100, 102, 124, and 312 were, with the exception of OTU-71, most abundant in the tunnel and little ocean pre-filtration tanks. OTU-71, in turn, was most abundant in the coral reef ditch tank. The water sample from the coral reef ditch also shared its most abundant OTUs with the sponge Stylissa carteri, namely, OTUs 17, 21, 37, and 70, classified as belonging to the Neptuniibacter genus and Arcobacteraceae family. These all had high BLAST sequence similarities (>99%) to organisms in GenBank obtained from seawater and a coral. OTUs 21 and 37 were both classified as belonging to the Malaciobacter genus (Phylum Campylobacteria). Another member of the Phylum Campylobacteria, OTU-79 classified as belonging to the Halarcobacter genus, was relatively abundant in Stylissa sp., in addition to Chalinula aff. milnei and Suberites aff. diversicolor, but only a minor component of water from the coral reef ditch. Halarcobacter and Malaciobacter are both genera within the Campylobacterales and are closely related to or synonymous with the genus Arcobacter. The genus Arcobacter has been extensively studied due to pathogenic activity in humans and animals (Ferreira et al., Reference Ferreira, Queiroz, Oleastro and Domingues2016). Members of the genus have also been detected in coastal marine waters and a number of invertebrates, among which the tropical reef sponge Spheciospongia solida (Cleary et al., Reference Cleary, Polónia, Reijnen, Berumen and de Voogd2020) and larval samples of the sponge Plakina cyanorosea (de Oliveira et al., Reference de Oliveira, Freitas-Silva, Sánchez-Robinet and Laport2020).

Figure 7. Relative abundances of the most abundant OTUs colour-coded according to prokaryotic phylum for: CinL, Cinachyrella sp.; SubR, Suberites aff. diversicolo; ChoF, Chondrosia aff. corticata; ChaT, Chalinula aff. milnei; ChrO, Chondrilla aff. australiensis; StyD, Stylissa carteri; TetO, Tethya aff. microstella; WatL, water lagoon; WatR, water reef gap; WatT, water tunnel; WatO, Little ocean pre-filtration tank; WatD, water coral reef ditch. The circle size of the OTU is proportional to the mean percentage of sequences per biotope as indicated by the symbol legend in the upper right corner of the figure below the phylum assignment legend. Note that for the sponges, the final letter of the code indicates the tank in which they were sampled, namely, L for lagoon, R for reef gap, T for tunnel, O for ocean pre-filtration tank, and D for coral reef ditch.
OTU-5, classified as belonging to the genus Lentibacter (Order Rhodobacterales), was recorded in all biotopes, but was most abundant in water from the lagoon and the sponge Cinachyrella sp. sampled from the lagoon tank. It had >99% BLAST sequence similarity to a microorganism obtained from coastal seawater (Supplementary data 4). The genus Lentibacter has previously been recorded at elevated levels in coastal waters and the microbiome of the jellyfish Rhizostoma pulmo (Stabili et al., Reference Stabili, Rizzo, Basso, Marzano, Fosso, Pesole and Piraino2020).
Several OTUs were mainly found in certain sponge species, e.g., Tethya aff. microstella (OTUs 75 and 271) and Chondrilla aff. australiensis (OTUs 130 and 169). Other dominant OTUs recorded in Tethya aff. microstella were OTUs 68, 190, and 333. These all had high BLAST sequence similarities (>99%) to microorganisms recorded in a coral, other sponge species, and seawater (Supplementary data 4). OTUs 68 and 333 were classified as belonging to the Sva0996 marine group. This group has been recorded in the bacterial communities of varying host sponge and coral species and is one of the few to exhibit a significant cophylogenetic signal (Moitinho-Silva et al., Reference Moitinho-Silva, Steinert, Nielsen, Hardoim, Wu, McCormack, López-Legentil, Marchant, Webster, Thomas and Hentschel2017; Steinert et al., Reference Steinert, Rohde, Janussen, Blaurock and Schupp2017; Dat et al., Reference Dat, Steinert, Cuc, Smidt and Sipkema2018; O'Brien et al., Reference O'Brien, Andreakis, Tan, Miller, Webster, Zhang and Bourne2021).
OTUs 46 and 52 were abundant in water from the tunnel and Chalinula aff. milnei sampled from the tunnel. These OTUs had high BLAST sequence similarities (>99%) to microorganisms found in seawater and marine seabed sediment. A large number of OTUs were recorded in sponges, but not in water (OTUs 2, 18, 19, 22, 23, 59, 68, 74, 84, 91, 96, 123, 155, 190, 206, 232, 245, 246, 249, 260, 285, 291, 318, and 333). Most of these had relatively high sequence similarities (>96%) to microorganisms mainly obtained from corals and sponges in GenBank (Supplementary data 4).
Relatively low sequence similarities were recorded for OTUs 2 (BLAST sequence similarity = 90%) and 84 (BLAST sequence similarity = 93%), classified as belonging to the BD72BR169 (Gammaproteobacteria) and P3OB-42 (Myxococcales) genera. Both OTUs were mainly found in Chondrosia aff. corticata and were related to microorganisms recorded in the soft coral Corallium rubrum and sediment, respectively (Supplementary data 4). Other OTUs mainly found in Chondrosia aff. corticata, but not water, were OTUs 59, 96, 123, 155, 206, 245, and 318. These were classified as belonging to a diverse amount of higher taxa including the BD2-11 terrestrial group, Nitrospirales, PAUC26f, PAUC34f, Nitrosococcales, and Oceanospirillales orders.
The BD2-11 terrestrial group (Phylum Gemmatimonadetes) includes members, which were previously found to be abundant in specimens of an aquarium cultured sponge, Haliclona cnidata (Schellenberg et al., 2020) and reef sponges from Vietnam (Dat et al., Reference Dat, Steinert, Cuc, Smidt and Sipkema2018). Rosales et al. (Reference Rosales, Miller, Williams, Traylor-Knowles, Young and Serrano2019) previously identified high abundances of P3OB-42 (Myxococcales) members in disease-free specimens of the coral Acropora palmata and suggested they may play a role in preventing disease. Myxococcales members can, furthermore, be facultative predators of other bacteria and may help to prevent disease by consuming pathogenic bacterial species (Welsh et al., Reference Welsh, Zaneveld, Rosales, Payet, Burkepile and Thurber2016). Nitrospira members are aerobic, chemolithoautotrophic bacteria. They are also all believed to be capable of nitrite oxidation and are often found in association with ammonia-oxidising bacteria or Archaea (Daims and Wagner, Reference Daims and Wagner2018). Certain members have, however, recently been found to be capable of complete nitrification (Daims et al., Reference Daims, Lebedeva, Pjevac, Han, Herbold, Albertsen, Jehmlich, Palatinszky and Vierheilig2015; van Kessel et al., Reference van Kessel, Speth, Albertsen, Nielsen, Op den Camp, Kartal, Jetten and Lücker2015). The high abundance of Nitrospira in Chondrosia aff. corticata suggests a symbiosis with potential relevance in the process of nitrite removal, which can be toxic to fish and other organisms.
OTUs 18, 19, and 22, classified as belonging to the Actinobacteria, NB1-j, and the genus Hormoscilla, respectively, were most abundant in Cinachyrella sp.. OTU-22, however, was only dominant in a single specimen (Bz004, where it accounted for 13,724 of 191,268 sequences), which clustered separately from the other Cinachyrella samples. OTU-22 was a rare component of all other sponge specimens varying from 13 to 142 sequences. Hormoscilla spongeliae, previously known as Oscillatoria spongeliae, has been associated with phototrophic sponges such as Lamellodysidea herbacea and appears to be unable to survive without its sponge host, which has thus far made it impossible to culture (Usher, Reference Usher2008; Schorn et al., Reference Schorn, Jordan, Podell, Blanton, Agarwal, Biggs, Allen and Moore2019). It is, however, known to produce halogenated compounds and polybrominated diphenyl ethers (PBDEs) with antimicrobial and antipredator properties (Flatt et al., Reference Flatt, Gautschi, Thacker, Musafija-Girt, Crews and Gerwick2005; Agarwal et al., Reference Agarwal, Blanton, Podell, Taton, Schorn, Busch, Lin, Schmidt, Jensen, Paul, Biggs, Golden, Allen and Moore2017). The species does not appear to be a normal symbiont of Cinachyrella species (Cleary and de Voogd, Reference Cleary and de Voogd2024). In Cleary and de Voogd (Reference Cleary and de Voogd2024), we showed that NB1-j members were relatively abundant in certain Cinachyrella specimens sampled from Martinique (C. alloclada) and Rodrigues, but were not observed or were only a rare component of other Cinachyrella species. In previous studies, we showed that dominant OTUs of Stylissa carteri were classified as NB1-j and were maintained across a large spatial scale (Cleary et al., Reference Cleary, de Voogd, Polónia, Freitas and Gomes2015; Reference Cleary, Polónia, Reijnen, Berumen and de Voogd2020; Reference Cleary, Polónia and de Voogd2021; Reference Cleary, Polónia, Swierts, Coelho, de Voogd and Gomes2022). In the present study, however, they were only a minor component of S. carteri.
OTU-23 (Order Rhodospirillales and Family Terasakiellaceae) dominated Suberites aff. diversicolor and accounted for almost 60% of the sequences. Specimens of Suberites diversicolor sampled from marine lakes in Berau and Papua, Indonesia were also dominated by two OTUs assigned to the Rhodospirillales (Ferreira et al., Reference Ferreira, Cleary, Coelho, Gomes, Huang, Polónia and de Voogd2020). This indicates that Suberites spp. appear to be able to maintain their dominant symbionts, even under highly modified environmental conditions. Metagenomic binning of sponge-associated vs free living Rhodospirillaceae revealed enrichment of genes required for the uptake and utilisation of organic sulphur compounds, particularly taurine, higher diversity and abundance of ABC transporters, and a distinct repertoire of genes involved in natural product biosynthesis, plasmid stability, cell detoxification and oxidative stress remediation (Karimi et al., Reference Karimi, Slaby, Soares, Blom, Hentschel and Costa2018). Rhodospirillaceae species can thrive under a broad range of environmental conditions and can function as chemotrophs, phototrophs, organotrophs and autotrophs (Esposti et al., Reference Esposti, Mentel, Martin and Sousa2019). Certain species can also use light in the infrared range enabling them to survive in darker environments (Solon et al., Reference Solon, Vimercati, Darcy, Arán, Porazinska, Dorador, Farías and Schmidt2018). Not much is known about the symbiotic role of Terasakiellaceae in sponges, but coral-associates of this family were suggested to be involved in the nitrogen metabolism (Weiler et al., Reference Weiler, Verhoeven and Dufour2018). Suberites aff. diversicolor was the only sponge species recorded in the reef gap tank.
In Figure 8, the results are shown for predicted metagenomic counts of six KEGG categories. The predicted relative gene counts of the biosynthesis of secondary metabolites, biosynthesis of antibiotics, Two-component system, and nitrogen metabolism categories were higher in the little ocean filtration and coral reef ditch tanks than in the other tanks. Interestingly, the little ocean filtration and coral reef ditch tanks had the highest and lowest nitrate levels, respectively (Supplementary data 1). Both the tunnel and little ocean tanks housed a variety of fish species, which contributed to the high nutrient levels in these tanks. In addition to this, effluent from the denitrification filters drains into the pre-filtration tanks, and this effluent had elevated nitrite levels (per. comm. MJ). Although nutrient levels were very low in the coral reef ditch tank, DOC levels were relatively high at 0.5 mg l−1.

Figure 8. Variation in the relative gene count abundance for (a) biosynthesis of secondary metabolites (metabolites), (b) biosynthesis of antibiotics (antibiotics), (c) carbon metabolism (carbon), (d) nitrogen metabolism (nitrogen), (e) sulphur metabolism (sulphur), and (f) two-component system (Two.component). The biotopes sampled were: CinL, Cinachyrella sp.; SubR, Suberites aff. diversicolor; ChoF, Chondrosia aff. corticata; ChaT, Chalinula aff. milnei; ChrO, Chondrilla aff. australiensis; StyD, Stylissa carteri; TetO, Tethya aff. microstella; WatL, water lagoon; WatR, water reef gap; WatT, water tunnel; WatO, Little ocean pre-filtration tank; WatD, water coral reef ditch. Note that for the sponges, the final letter of the code indicates the tank in which they were sampled, namely, L for lagoon, R for reef gap, T for tunnel, O for ocean pre-filtration tank, and D for coral reef ditch.
Among sponges, the single sponge species residing in the coral reef ditch, Stylissa carteri, also had the highest relative gene counts of the biosynthesis of secondary metabolites, nitrogen metabolism, and Two-component system categories and the lowest relative gene counts of the carbon metabolism category. The coral reef ditch water sample and S. carteri both housed abundant OTUs assigned to the genus Neptuniibacter in the Nitrinocolaceae family. Neptuniibacter members have the capacity to assimilate taurine–nitrogen and have been implicated in carbazole degradation (Diéguez et al., Reference Diéguez, Balboa, Magnesen and Romalde2017). A putative Neptuniibacter member was also part of a phenanthrene-degrading community following an oil spill and appeared to be a sulphur oxidiser capable of degrading sulphur-containing oil components. Categories including nitrate and nitrite ammonification, dissimilatory nitrite reductase and denitrification were also detected in the genome of a Neptuniibacter member (Dombrowski et al., Reference Dombrowski, Donaho, Gutierrez, Seitz, Teske and Baker2016).
Sponge specimens residing in the lagoon and reef gap tanks had higher predicted gene counts of the biosynthesis of antibiotics category than sponges in the other tanks. Gene counts for the carbon metabolism category were higher in water from the lagoon and reef gap tanks and the nitrogen metabolism, sulphur metabolism, and two-component system categories in water from the other tanks. Among sponges, the most marked difference was the predicted enrichment of the Sulphur metabolism category in sponges residing in the tunnel, little ocean pre-filtration tank, and coral reef ditch tanks compared to the lagoon and reef gap tanks.
We identified 31 potential pathogens, given the limitations of short 16S RNA gene sequences, and 20 potential beneficial organisms in our data set (Figure 9 and Supplementary data 5). The OTUs in question had ≥ 99% sequence similarity to known pathogenic organisms and potentially beneficial organisms. Overall, the percentage of potential pathogens varied from 0.03% in one sample of Chondrilla aff. australiensis to 7.48% in water from the reef gap. The potential pathogens included OTU-355, related to Photobacterium damselae, and OTU-220, related to Vibrio harveyi. OTU-355 was most abundant in water from the lagoon and reef gap tanks where it accounted for 1.05% and 4.48% of all sequences, respectively. It was much less abundant in all other biotopes only reaching 0.3% of sequences in Cinachyrella sp. and less than that in the remaining sponge and water samples. OTU-220 was also most abundant in water from the lagoon and reef gap tanks; in the lagoon it accounted for 1.46% of all sequences. It was much less abundant in all other biotopes. Other less abundant potential pathogens included V. vulnificus (OTU-660), Mycobacterium fortuitum (OTU-3320), M. ulcerans (OTU-16858), Pseudoalteromonas undina (OTUs 364, 1210, 1353, 5851), Aeromonas hydrophila (OTU-11214), Pseudomonas plecoglossicida (OTU-3304), and Clostridium botulinum (OTU-20656) among others. Photobacterium damselae and V. harveyi are well known pathogens of marine animals and are responsible for disease outbreaks in aquaculture systems (Austin and Zhang, Reference Austin and Zhang2006; Rivas et al., Reference Rivas, Lemos and Osorio2013). Interestingly, the elevated predicted pathogen load in water from the lagoon and reef gap tanks coincided with enrichment of the biosynthesis of antibiotics category in sponges from these tanks suggesting that the sponges may help to limit pathogen growth although this needs further verification. The relatively high potential pathogen loads of the lagoon and reef gap tanks also requires additional investigation. It should be noted that following sampling, a UV-C filter was added to the filtration system of the lagoon tank.

Figure 9. Relative abundances of potential pathogens and potential beneficial microorganisms in: CinL, Cinachyrella sp.; SubR, Suberites aff. diversicolor; ChoF, Chondrosia aff. corticata; ChaT, Chalinula aff. milnei; ChrO, Chondrilla aff. australiensis; StyD, Stylissa carteri; TetO, Tethya aff. microstella; WatL, water lagoon; WatR, water reef gap; WatT, water tunnel; WatO, Little ocean pre-filtration tank; WatD, water coral reef ditch. Note that for the sponges, the final letter of the code indicates the tank in which they were sampled, namely, L for lagoon, R for reef gap, T for tunnel, O for ocean pre-filtration tank, and D for coral reef ditch.
OTUs in the present study were related to a number of potentially beneficial bacteria including Pseudovibrio denitrificans (OTU-878), Alteromonas macleodii (OTU-92), Ruegeria mobilis (OTU-1754), Phaeobacter piscinae (OTU-3543), and Pseudoalteromonas luteoviolacea (OTU-856). OTU-878 was most abundant in Chalinula aff. milnei and accounted for almost 1% of all sequences on average. The percentage of potentially beneficial bacteria among samples, furthermore, varied from 0.00% in S. carteri to 3.11% for one sample of Chalinula aff. milnei. Pseudovibrio denitrificans is a heterotrophic and facultatively anaerobic marine bacterial species described from specimens collected in seawater from Taiwan (Shieh et al., Reference Shieh, Lin and Jean2004). Members of the genus play a role in denitrification and can use various terminal electron acceptors including nitrate, nitrite or nitrous oxide (Shieh et al., Reference Shieh, Lin and Jean2004). In a shrimp (Penaeus vannamei) aquaculture setting, Pseudovibrio members isolated from the sponge species Aplysina gerardogreeni were shown to inhibit the growth of pathogenic Vibrio species (Domínguez-Borbor et al., Reference Domínguez-Borbor, Ardiles, Bermeo, Bolívar-Alvarado, Tomalá, Sonnenholzner and Rodríguez2019). Nicacio et al. (Reference Nicacio, Ióca, Fróes, Leomil, Appolinario, Thompson, Thompson, Ferreira, Williams, Andersen, Eustaquio and Berlinck2017) also showed that Pseudovibrio denitrificans Ab134 obtained from the sponge Arenosclera brasiliensis produced a range of bromotyrosine-derived alkaloids (Nicacio et al., Reference Nicacio, Ióca, Fróes, Leomil, Appolinario, Thompson, Thompson, Ferreira, Williams, Andersen, Eustaquio and Berlinck2017).
OTU-92 (genus Alteromonas; Gammaproteobacteria) was less abundant than OTU-878, but was observed in all of the water samples. It had 100% sequence similarity to an organism identified as Alteromonas macleodii. A widespread copiotroph found in temperate and tropical surface waters, it has been shown to be able to use the complete pool of labile DOM-derived compounds (Pedler et al., Reference Pedler, Aluwihare and Azam2014). Certain strains are also known to associate with Prochlorococcus species by means of enhanced phenol degradation (López-Pérez et al., Reference López-Pérez, Gonzaga, Martin-Cuadrado, Onyshchenko, Ghavidel, Ghai and Rodriguez-Valera2012). They also appear sensitive to iron limitation (Fourquez et al., Reference Fourquez, Devez, Schaumann, Gueneugues, Jouenne, Obernosterer and Blain2014). In terms of benefits, A. macleodii was shown to enhance growth of the microalga Isochrysis galbana, among the most important of bait microalgae in aquaculture (Cao et al., Reference Cao, Wang, Wu, Kong, Lin, Ling, Xu, Ma, Zhang, Zhou, Yan and Xu2021). It has also been used as a probiotic in mussel aquaculture (Kesarcodi-Watson et al., Reference Kesarcodi-Watson, Kaspar, Lategan and Gibson2010).
Conclusion
The results of the present study echo those of previous studies in natural settings, which highlighted the distinct nature of sponge-associated prokaryotic communities in comparison to those found in water (Lee et al., Reference Lee, Wang, Yang, Lafi, Al-Suwailem and Qian2011; Reveillaud et al., Reference Reveillaud, Maignien, Eren, Huber, Apprill, Sogin and Vanreusel2014; Cleary et al., Reference Cleary, Polónia, Becking, de Voogd, Purwanto, Gomes and Gomes2018b). The sponge fauna of Burgers' Zoo Ocean aquarium consisted of a variety of sponges with different life strategies and distinct prokaryotic communities. Sponges in the present study also hosted microorganisms potentially involved in nitrification, denitrification, sulphur oxidation, and antibiotic biosynthesis. Due to their water filtering capacities, they may play an important role in maintaining a healthy aquarium environment. Taken together, our study shows that diverse sponge-associated and bacterioplankton communities have colonised Burgers' Zoo Ocean aquarium. The sponge-associated prokaryotic communities included OTUs with affinity to organisms obtained from other sponges in the natural environment, but their composition also appeared to vary with water chemical parameters and bacterial composition of the water in which they were collected suggesting an important effect of the aquarium environment.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0025315424000602.
Data availability
The DNA sequences generated in this study can be downloaded from NCBI BioProject Id: PRJNA715763.
Acknowledgements
We wish to thank the Burgers' Ocean team for logistical support, sample collecting, and laboratory assistance.
Author contributions (for papers with two or more authors)
DFRC and NJdV wrote the main part of the manuscript. DFRC analysed the data. NJdV identified the sponge specimens. MJ provided logistical support. DFRC, NJdV, MJ, TMS, and NCMG all contributed to later stages of the manuscript.
Financial support
This work was supported by European Funds through COMPETE [FCOMP-01-0124-FEDER-008657] and by National Funds through the Portuguese Foundation for Science and Technology (FCT) within the LESS CORAL [PTDC/AAC-AMB/115304/2009] and Ecotech-Sponge (PTDC/BIAMIC/6473/2014 – POCI-01-0145-FEDER-016531) projects. This work is also part of the research programme NWO-VIDI with project number 16.161.301, which is (partly) financed by the Netherlands Organisation for Scientific Research (NWO). We acknowledge financial support to CESAM from FCT/MCTES (UIDP/50017/2020 + UIDB/50017/2020 + LA/P/0094/2020), through national funds.
Competing interest
The authors declare that there were no conflicts of interest.
Ethical standards
Ethical standards were maintained during the course of the study.