INTRODUCTION
Streptococcus pneumoniae is a major cause of morbidity and mortality worldwide and causes a diverse range of diseases including pneumonia, meningitis, bacteraemia and otitis media. The polysaccharide capsule plays a major role in the virulence of the organism and there are more than 90 capsular serotypes [Reference Park1]. However, the distribution of serotypes varies between countries and within the United Kingdom, invasive pneumococcal disease (IPD) is associated with a smaller number of serotypes [Reference Ihekweazu2]. Children aged <5 years and the elderly are particularly at risk of developing IPD. In the United Kingdom two vaccines have been licensed to prevent IPD. A 23-valent plain pneumococcal polysaccharide vaccine (PPV) which stimulates a T-cell-independent immune response in adults and in children aged >2 years, but is poorly immunogenic in younger children. The antibody response in children aged <2 years is improved by the use of a protein-conjugated pneumococcal vaccine (PCV) which induces a T-cell-dependent response [Reference Goldblatt, Siber, Klugman and Mäkelä3]. PPV immunization of elderly patients was introduced in England and Wales in three phases: in 2003 PPV was offered to all patients aged ⩾80 years; in 2004 it was offered to all aged ⩾75 years; and from 2005 onward all aged ⩾65 years. The efficacy of PPV is controversial [Reference Conaty4–Reference Vila-Corcoles7]. In September 2006, a 7-valent PCV was incorporated into the routine infant immunization schedule in England and Wales, with a reduced schedule of 2, 4 and 13 months.
Surveillance of IPD and molecular characterization of isolates causing IPD are fundamental to understanding the impact of pneumococcal vaccines and to assessing whether changes in the genetic structure of the pneumococcal population occur following a universal immunization programme. Precise genetic analyses that may reflect the genetic relatedness of the isolates and the evolutionary biology of pneumococci are required to detect potential changes in the genetic structure of the pneumococcal population. Assessment of the genetic characteristics of pneumococci causing IPD in the elderly prior to the introduction of the PCV is required to detect any changes caused by indirect herd effects. In this paper the serotypes and genotypes of pneumococci causing IPD in the elderly prior to introduction of PCV are described.
MATERIAL AND METHODS
All isolates from patients targeted by the first year of the PPV programme were selected from the collection of the Respiratory and Systemic Infection Laboratory (RSIL), the reference laboratory for serotyping pneumococci in England and Wales. The criteria for selection were as follows: all invasive (blood or CSF) isolates regardless of serotype from patients aged ⩾80 years collected during winter 2003/2004 (1 November 2003 to 30 April 2004) in England and Wales. As part of the HPA surveillance of IPD in England and Wales, details of any PPV immunization for these patients were obtained from the general practitioner. PPV failure was defined as an episode of IPD caused by a PPV serotype occurring during the period of 1 month to 2 years after PPV immunisation. All isolates were serotyped by an agglutination method using factor sera from the Statens Serum Institute, Denmark and further characterized by multilocus sequence typing (MLST) [Reference Enright and Spratt8]. Briefly, 10–20 colonies from an overnight blood agar plate were boiled for 20 min in saline solution (NaCl 9‰ w/v) and cell debris were separated from DNA extracts by centrifugation at 3000 rpm for 10 min. The supernatant was stored at −20°C before amplification. Fragments of the seven housekeeping genes aroE, gdh, gki, recP, spi, xpt, ddl were amplified by PCR using Invitrogen Taq polymerase and PCR buffer and purified PCR products were directly sequenced at MWG Biotech (Ebersberg, Germany). Allele numbers and sequence types (ST) were assigned as described by Platt et al. [Reference Platt9] using the BioNumerics version 4.0 platform (Applied Maths, Sint-Martens-Latem, Belgium) with reference to the MLST database (http://spneumoniae.mlst.net). When a new association of ST and serotype was observed, serotyping was repeated to confirm the serotype. Genetic relatedness between isolates was determined using eBURST V3 algorithm [Reference Feil10]. STs obtained from the RSIL collection were analysed against all STs reported in the MLST database in order to cluster the population into clonal complexes (CC). CC was defined using the stringent group definition (6/7 shared alleles) and named referring to the predicted founder if any (i.e. CC156 referred to STs having ST156 as predicted founder) or referring to the eBURST group number if no founder were predicted (i.e. gr103 referred to eBURST group 103). Genetic diversity based on ST and CC was estimated by calculating the Hunter–Gaston Index [Reference Hunter and Gaston11] using the V-DICE application (available at http://www.hpa-bioinformatics.org.uk).
RESULTS
A total of 542 invasive pneumococci isolated during the winter of 2003/2004 from patients aged ⩾80 years were examined. A total of 32 serotypes were identified among these isolates (Table 1). IPD caused by the capsular types included in PCV or PPV accounted for 55·5% and 91·9%, respectively, of the total of cases. Serotype 14 was the predominant serotype in this collection. The 10 most common serotypes accounted for 75·6% of all isolates and comprised serotypes 14 (18·5%), 23F (11·8%), 9V (9·2%), 3 (6·1%), 6B (5·9%), 22F (5·4%), 4 (5·0%), 19A (4·8%), 8 (4·6%), and 6A (4·4%).
Table 1. Phenotypic and genotypic characterization of pneumococci causing invasive diseases in elderly patients (England & Wales, winter 2003–2004)
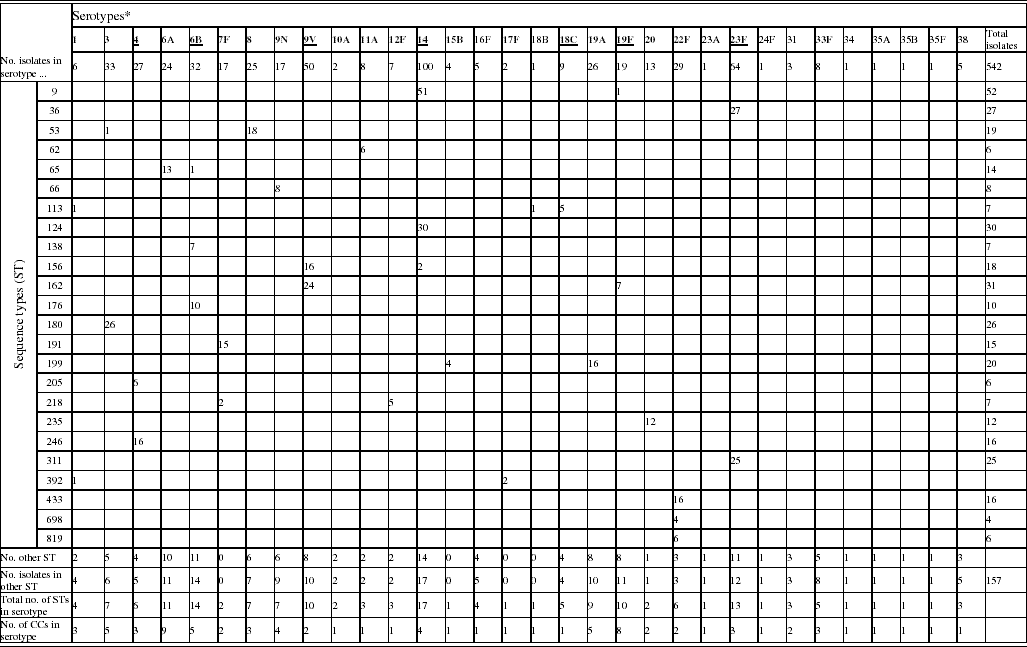
* Serotypes underlined are included in pneumococcal conjugate vaccine (PCV), serotypes in bold are included in polysaccharide pneumococcal vaccine (PPV).
Of the 542 isolates, MLST analyses identified 144 STs. The Hunter–Gaston Diversity Index (HGDI) was high (0·97). ST9 represented 9·5% of the total STs and was predominant in the bacterial population followed by ST162 (5·7%), ST124 (5·5%), ST36 (5·0%), ST180 (4·8%) and ST311 (4·6%) (Table 1). These six major STs accounted for 35·2% of all isolates. A strong association between ST and serotype was observed for the common STs. Ninety-two STs were identified once only. At the time of the study, 55 STs were novel types in the international MLST database. The genetic relatedness of the pneumococci was assessed by eBURST analysis. STs were clustered into 27 CCs and 35 singletons (Fig. 1).
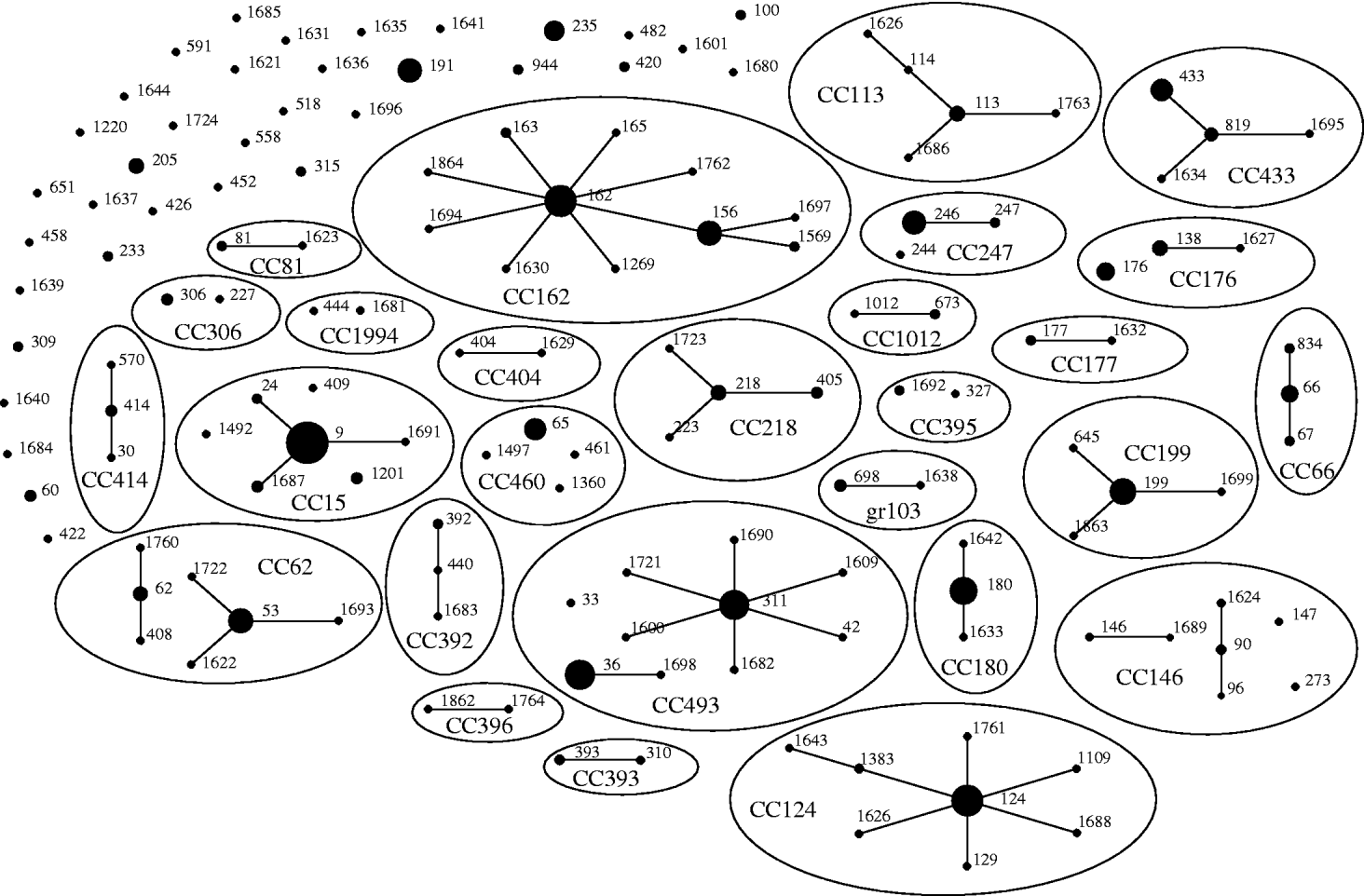
Fig. 1. Genetic relatedness of 542 isolates of S. pneumoniae causing invasive disease in elderly patients in the United Kingdom. Clonal complexes (CCs) and singletons were determined by eBURST analysis using all allelic profiles reported in the MLST database. Only the 144 sequence types (STs; black dots) detected in this study were represented on the eBURST diagram. The sizes of the dots correspond to the number of isolates sharing the same ST. Within CCs (elliptic shapes) missing links (STs present in the MLST database but not found in the present study) were not represented. CC names refer to the predicted founder if any (i.e. CC156 referred to STs having ST156 as predicted founder) or refer to the eBURST group number if no founder were predicted (i.e. gr103 referred to eBURST group 103).
The HGDI based on ST as variable (HGIST) by serotype ranged from 0·233 (7F) to 0·882 (19F) (Table 2). The HGDI calculated using CC rather than ST as variable (HGICC) was, as expected, lower for almost all serotypes as eBURST reduces the diversity by clustering closely related STs into CCs. Difference between HGIST and HGICC was marked for serotypes 9V, 18C, and 23F (HGIST>0·65 and HGICC<0·15) but was close to null for serotypes 6A and 19F for which values of both HGIST and HGICC were high (>0·7). For serotype 7F the value of both indices was equal.
Table 2. Hunter–Gaston indices of the major serotypes causing invasive pneumococcal disease in elderly patients (England & Wales, winter 2003–2004) using as variable sequence types or clonal complexes calculated by eBURST
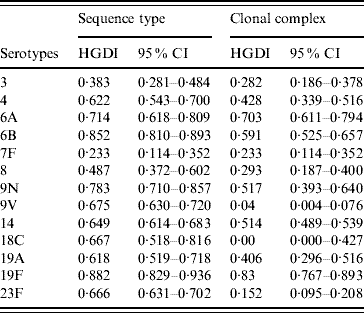
HGDI, Hunter–Gaston Diversity Index; CI, confidence interval.
Distinct serotypes were observed in nine STs: ST9, ST53, ST65, ST113, ST156, ST162, ST199, ST218 and ST392 (Table 1). The number of different serotypes within these STs was low (1 or 2 per ST). ST113 which is generally associated with serotype 18C was found to be associated with serotypes 1 and 18B. Within each of the major STs the proportion of serotype variants was always low in comparison to the commonest serotype.
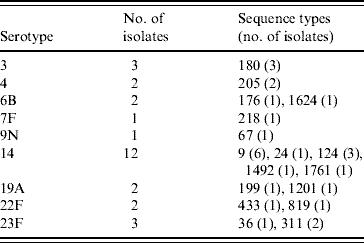
The PPV immunization history was established for 378 patients: 231 patients had never received PPV or received PPV <1 month prior to the infection, 40 patients had received PPV in the 2 years preceding the episode of IPD, and 107 patients had received PPV >2 years prior to the IPD. Twenty-eight PPV failures were identified amongst the 40 patients that had been vaccinated within 2 years prior to infection. Vaccine failures were associated with nine serotypes and MLST identified 17 STs (Table 3); ST1761, belonging to CC124, associated with serotype 14 and ST1624, belonging to CC146, associated with serotype 6B were new entrants in the MLST database. The other STs had already been reported to the international database.
Table 3. Genetic characterization of pneumococci causing pneumococcal polysaccharide vaccine failure in elderly patients vaccinated within 2 years prior to infection (England & Wales, winter 2003–2004)
DISCUSSION
In this study, serotyping and MLST genotyping were used to characterize invasive pneumococci collected from elderly patients aged ⩾80 years in England and Wales. During the period November 2003–April 2004, pneumococci of serotypes 14, 23F and 9V accounted for >40% of the isolates causing IPD. Genetic diversity within the pneumococcal population causing IPD in elderly patients was high. Of 144 distinct STs, ST9 was the most prevalent clone. ST9 is recognized as the international erythromycin-resistant clone England14-9. Antibiotic susceptibility testing on isolates of ST9 collected since 2002 in all age groups have shown that almost all of these isolates were erythromycin non-susceptible (data not shown). It may be that usage of antibiotics contributes, at least partially, to the prevalence of drug- resistant clones as suggested by Feikin et al. [Reference Feikin12]. In the United States, the recent expansion of clones of serotype 19A following the introduction of the PCV was caused by the emergence/expansion of penicillin-resistant clones [Reference Moore13]. Other possible explanations for the prevalence of certain serotypes in the elderly has been discussed previously [Reference Feikin12] and may include, the effect of the senescence on the immune system and contacts with children, in addition to the selective pressure of antibiotics.
The predominant serotypes observed in the present study comprised genetically diverse clones as demonstrated by previous studies in the United Kingdom [Reference Clarke14, Reference Jefferies15]. Within a single serotype, genetic variability may have arisen largely by divergence from ancestral clones through evolutionary biology. However our data suggest that pneumococcal serotypes causing IPD could be grouped into two classes. The first class includes serotypes (e.g. 6A and 19F and to a lesser extent 6B, 9N and 14) which are shared by numerous STs related to diverse pneumococcal lineages. The second group includes serotypes (e.g. 3, 7F, 8, 9V, 18C and 23F) which contains STs related to single or very few pneumococcal lineages. Serotypes of the first class were among the most common serotypes detected in UK carriage studies in children [Reference Hussain16–Reference Sandgren18] while serotypes of class 2 were rarely carried, except for serotype 23F. Pneumococci have the ability to exchange their capsular type through horizontal recombination of pneumococcal DNA surrounding the capsular loci by natural transformation [Reference Coffey19]. Capsular switching (CS) most probably occurs during nasopharyngeal carriage where intermixing of clones expressing distinct types of capsular polysaccharides is possible. The propensity of certain pneumococci to be carried and to undergo CS may have contributed to the differential diversity of pneumococcal lineages between serotypes.
Serotype 6A was one of the more genetically diverse serotypes. However, it may that such diversity was overestimated by the presence of isolates expressing 6C capsular polysaccharides [Reference Park1] as distinction between the two subtypes (6A and 6C) was not included in the reference typing at the time of this study.
Among invasive isolates of serotype 23F only three pneumococcal lineages have been identified (CC81, CC392, and CC439). Serotype 23F is frequently found in healthy carriers [Reference Jefferies15–Reference Sleeman17] and can colonize the nasopharynx for several weeks or months [Reference Sleeman17]. It may be that either 23F pneumococci are less prone to CS or only a few clones are able to cause IPD in elderly patients. A combination of properties associated with particular clones and capsular serotypes is likely to determine the potential of pneumococci to cause invasive diseases, their manifestations and severity [Reference Sandgren18, Reference Brueggemann20].
There was no direct evidence of CS but combining the MLST and serotype, data suggest that CS may have occurred in nine STs in the past. Amongst clones that have undergone CS, the proportion of serotype variants was always lower in comparison to the main serotype within that clone. CS may have occurred in the very recent past and thus the emerging serotype variants have not had time to disseminate within the population. Alternatively, the serotype variants derived from CS may be less virulent/invasive and, therefore, spread at a slower rate in the elderly population than the original clones.
Our data indicated that 55% of IPD in the elderly were due to the seven serotypes included in PCV. This suggested that the recent introduction of PCV in UK childhood immunization could have a substantial impact on the distribution of the serotypes causing IPD in the elderly through an indirect herd effect as has been observed in the United States [Reference Lexau21]. Beneficial indirect herd effects of PCV could be limited by the emergence of clones that switch their capsule type from a type included in PCV to a type not included in PCV via CS. In our collection we found a serotype 1 variant of the virulent clone ST113 which is commonly associated with serotype 18C (according to the MLST database; last accessed 28 September 2008). It suggests that virulent clones that could escape the selective pressure of PCV existed prior to introduction of the vaccine in the United Kingdom. In the United States, changes in serotype distribution have been observed since the introduction of PCV [Reference Whitney22–Reference Jacobs26] resulting in substantial changes to the genetic structure of the pneumococcal population [Reference Moore13, Reference Jacobs26, Reference Beall27]. Changes were mainly due to the expansion/emergence of drug-resistant clones [Reference Moore13, Reference Pai28–Reference Messina31] but were also due to the expansion of a clone that had undergone CS from PCV serotype to non-PCV serotype [Reference Moore13, Reference Brueggemann32].
Prior to the introduction of the routine PPV vaccination programme for the elderly PPV was recommended for adults at high risk of developing IPD and the uptake for adults aged ⩾80 years was estimated to be 36% [Reference Noakes33]. Between August 2003 and March 2004, the additional vaccine uptake for elderly persons aged ⩾80 years was 26·1% [34]. Apparent PPV failures were observed in 28 patients. Two cases were vaccine failures caused by novel STs: ST1624 (single locus variant of the multidrug-resistant clone Spain6B-2) and ST1761. However, data on these two novel STs are too limited to conclude that vaccine failures were caused by clones emerging under PPV selection pressure.
Surveillance of IPD in the United Kingdom will continue in order to assess the effects of the vaccination programmes for the elderly and children on the genetic characteristics of isolates causing IPD in the elderly as well as in children. By targeting the reservoir of the pathogen, it is anticipated that PCV immunization of children should exert a more selective pressure on pneumococci than PPV immunization of older persons.
ACKNOWLEDGEMENTS
This study was funded by the UK Department of Health. Thanks are due to Nick Andrews from the Statistics, Modelling and Bioinformatics Department – CfI, HPA London, and Usha Gungabissoon, Rashmi Malkani, Elizabeth Miller and Pauline Waight from the Immunization Department – CfI, HPA London for collecting patients' vaccination history data. This publication made use of the Multi Locus Sequence Typing website (http://www.mlst.net) at Imperial College London, developed by David Aanensen and funded by the Wellcome Trust.
DECLARATION OF INTEREST
None.






