More than 180 nontuberculous mycobacteria (NTM) in natural soil and water as environmental resident bacteria have been reported.Reference Phillips and von Reyn1 NTM occasionally enter the human body through the respiratory or digestive tracts. Some NTM species, including slow-growing species, such as Mycobacterium avium/M. intracellulare [M. avium complex (MAC)] and M. kansasii, and rapidly growing species, such as M. abscessus, cause diseases in humans.Reference Phillips and von Reyn1 M. lentiflavum is a slow-growing NTM that was first isolated in 1996.Reference Springer, Wu and Bodmer2 It rarely causes diseases in healthy humans and is detected in soil and water, similar to other NTM.Reference Tsitko, Rahkila and Priha3,Reference Marshall, Carter and Torbey4 However, infections caused by M. lentiflavum can occur in immunocompromised hosts, such as those with chronic respiratory disease, leukemia, those undergoing dialysis, or those with acquired immunodeficiency syndrome.Reference Tortoli, Bartoloni and Erba5,Reference Yagi, Morimoto and Ishii6
NTM detection in clinical samples, such as sputum, does not always imply an infection, and the possibility of patient colonization or environmental contamination needs to be considered.Reference Miyamoto, Yamaguchi and Sasatsu7 For NTM diagnosis, mycobacterial culture of bronchoalveolar lavage fluid (BALF) samples obtained using a bronchoscope is considered to be more diagnostically valuable than mycobacterial culture of sputum samples.Reference Griffith, Aksamit and Brown-Elliott8 Pseudo-outbreaks of NTM caused by bronchoscope contamination have been reported.Reference Takajo, Iwao and Aratake9–Reference Guimaraes, Chimara and do Prado12 To our knowledge, no pseudo-outbreak with M. lentiflavum has been reported. We report the genomic and epidemiologic investigation into a pseudo-outbreak caused by M. lentiflavum in a Japanese hospital.
Methods
Study oversight
The present study was conducted under the approval of the Research Ethics Committee of Kushiro City General Hospital (no. 2021-10) and in accordance with the provisions of the Declaration of Helsinki. Informed consent was obtained from all the patients by the opt-out method using the hospital web page.
NTM isolation and identification
NTM-positive clinical samples collected from patients between May 2020 and April 2021 at Kushiro City General Hospital in Japan were analyzed. We reviewed the medical records of patients whose isolates were identified as M. lentiflavum. We investigated the number and species of all mycobacterial isolates at our hospital from January 2018 to April 2021.
As a routine clinical procedure for mycobacterial screening in the hospital, the collected samples were tested by both acid-fast staining and culture. The samples collected from patients were first tested by Ziehl–Neelsen staining followed by liquid culture (Bactec MGIT 960 System, Nippon Becton, Dickinson, Tokyo, Japan) and cell culture in the Ogawa medium (Kyokuto Pharmaceutical Industrial, Tokyo, Japan). Positive cultures were first tested for M. tuberculosis and MAC by nucleic acid amplification test using TRCReady (Tosoh Bioscience, Tokyo, Japan).
No M. lentiflavum isolates were identified before May 2018; in retrospect, that absence reflects limitations of the systems in use for identifying the species of NTM isolates. Following introduction in the hospital microbiology laboratory of matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy (MALDI-TOF MS microflex, Bruker Japan, Kanagawa, Japan) for identification of NTM, M. lentiflavum isolates were reported.
All samples were spotted on a MALDI target plate. Altogether, 24 MALDI-TOF spectra were acquired per sample using the automated functionality of the flexControl 3.0 software (Bruker Daltonik, Billerica, MA). The raw spectra were quality controlled and used to generate a main spectrum (MSP) with the MALDI Biotyper 4.0 software (Bruker Daltonik) with default settings. The created MSPs were matched against the Mycobacteria Library version 4.0 (Bruker Daltonik) with 880 references. In this study, bacterial identification was also performed by ANI analysis as described previously.Reference Yoon, Ha, Lim, Kwon and Chun13 We used de novo assembled genomes of NTM isolates obtained from WGS as described in “Molecular analyses of M. lentiflavum” for ANI analysis.
We used M. lentiflavum ATCC51985 as the reference genome.Reference Niobe, Bebear, Clerc, Pellegrin, Bebear and Maugein14
Hospital investigation
Bronchoscopy for outpatients is performed in one room of the respiratory medicine outpatient clinic, and, before the test, bronchoscope leak testing is performed using tap water. After the examination is completed, the equipment used is cleansed using an automatic washer in the next room and returned to the storage rack in the respiratory medicine outpatient clinic. The hospital has 5 automatic washers; they are also used to wash gastrointestinal endoscopes. These units use 6% ethaneperoxoic acid for cleaning instruments, which is then checked to maintain a concentration of at least 0.2% daily. From June 2020 to January 2021, the concentration did not fall below 0.2%. Bronchoscopies for inpatients are performed with the same bronchoscopes in other areas, and the instruments are cleaned using the same automatic washers. The difference between outpatient and inpatient procedures is the origin of tap water for the bronchoscope leak test.
Sputum samples, which are collected in the respiratory medicine and the emergency medicine, are carried directly to the bacteriological laboratory in the hospital. When patients are not able to produce sputum, they inhale distilled water by nebulizer.
Environmental investigation
Kushiro City General Hospital was built at its current location in 1984 and has undergone several expansions. Its water supply system was originally a single system, but a dual water supply system was created when the building was renovated in 2008.
Water is supplied from Kushiro City, Hokkaido, via waterworks and is first stored in an underground tank. Originally, only 1 system was used to pump water from the underground tank to a tank placed on the roof, and water was distributed to various hospital areas by gravity. However, due to the hospital expansions, a new water supply system utilizing a pump was created, which distributes water directly from the underground tank. The respiratory medicine department is among the areas supplied by the new water system, whereas the emergency department is supplied by the old water system. The supplied water comes from the Kushiro River, which flows out from Lake Kussharo, and the water quality is based on the water quality control standards of Japan.
Rinse-water samples from 4 bronchoscopes, water samples from automated endoscope washers, and water samples directly from the water supply in the respiratory medicine and the emergency medicine, which include the water storage tank, were collected. The samples were processed and inoculated into liquid media. All Mycobacterium spp cultured were identified to the species level.
Given that the water supply system can be a source of NTM, we collected a total of 52 environmental samples (Supplementary Appendix Table S1 online). The precipitates of the samples were then tested by acid-fast staining and culture. When positive results were obtained, the nucleic acid amplification test, MALDI-TOF MS, and average nucleotide identity (ANI) analysis were performed to identify the bacteria.
Molecular analyses of M. lentiflavum
To identify M. lentiflavum, we used clinical isolates collected from May 2020 to April 2021 and isolates from the environmental samples collected on February 26, 2021.
Genomic DNA was extracted from the bacteria by using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany). A DNA library for WGS was prepared using the Nextera XT DNA library preparation kit (Illumina, CA) according to the manufacturer’s instructions. The libraries were sequenced on a MiSeq system sequencer (Illumina) with 300-bp paired-end reads according to the manufacturer’s protocol. The trimming and de novo genome assembly were performed by using the CLC Genomics Workbench (Qiagen). The multilocus sequence typing (MLST) was performed using the software provided by the Center for Genomic Epidemiology (https://cge.cbs.dtu.dk/services/MLST/ for Mycobacteria spp).Reference Jolley, Bray and Maiden15 Core genome and accessory genome analyses were performed using Roary: the Pan Genome Pipeline version 3.11.2 as the default software.Reference Page, Cummins and Hunt16
The number of single-nucleotide polymorphisms (SNPs) was calculated by comparison with patient 1 and patient 2 assembly genomes. The trimmed nucleotide sequence (contigs) of each isolate was mapped on the patient 1 or patient 2 genomes, respectively. Then, CLC Genomic Workbench basic variant detection was used for the SNP identification as a default parameter (ploidy was 2, minimum coverage, count, and frequency were 10, 5, and 50%, respectively).
Results
Eligible patients and environmental samples
Altogether, 100 cases were culture-positive among the samples obtained from May 2020 to April 2021. Of these, 22 samples were positive for M. lentiflavum according to the MALDI-TOF MS results (Table 1). All patient specimens were provided by the respiratory medicine department. The sputum specimens from 8 patients and bronchoscope-derived specimens from 14 patients were collected from the respiratory (n = 19) and emergency (n = 3) departments. Two patients (patient 13, with interstitial pneumonia being treated with steroids and patient 16, undergoing chemotherapy for a solid tumor malignancy) were considered susceptible to infection based on their underlying conditions. Also, 9 patients were suspected of having mycobacterial infections.
Table 1. Characteristics of the Eligible Patients

Note. BALF, bronchoalveolar lavage fluid.
a The type of samples.
b Areas where samples were collected. Respiratory, respiratory outpatient; Emergency, emergency outpatient.
No patients tested positive again for M. lentiflavum, although multiple subsequent bacteriological examinations were performed. Before the genetic analysis was performed, the excessive detection of M. lentiflavum had been already a clinical problem. Since we considered the frequency of M. lentiflavum isolation itself to be basically low, we suspected the possibility of contamination. Subsequently, only 1 of these cases was treated for M. lentiflavum. During follow-up, no progression of disease that could be considered an exacerbation of M. lentiflavum was noted. Clinically, we strongly suspected these cases as contamination.
Patient 10 was suspected of having an NTM infection owing to the presence of lung nodules. She underwent bronchoscopy, and M. lentiflavum grew from her BALF. Because the excessive detection of M. lentiflavum had not been shared among attending physicians at the time her culture was detected, she was diagnosed with a M. lentiflavum infection and was treated with rifampicin, ethambutol, and clarithromycin. These medications were discontinued on day 2 because she developed a drug fever. She was monitored thereafter, but no worsening of disease was observed and no subsequent M. lentiflavum cultures were positive.
Chart review revealed that most patients lived in the city where the hospital was located. Moreover, 3 patients lived >50 km away, and 1 patient whose isolate was genetically similar lived >150 km from the hospital. No patients had consanguineous relationships. All patients were outpatients, and the samples were collected from the respiratory medicine department (n = 19) and the emergency department (n = 3).
Among the environmental culture specimens, 1 faucet sample from the respiratory medicine outpatient department, submitted in February 2021, grew M. lentiflavum. The tap water was used for the bronchoscope leak testing, nebulizer cleaning for sputum examination, and staff hand washing.
Genetic analysis
All NTM isolates were identified as M. lentiflavum by ANI analysis (ANI value was > 99.2%). Altogether, 23 specimens, including 22 patient-derived specimens and 1 environmental culture specimen, were examined by WGS analysis (Table 2). The results showed sampling errors due to the poor extraction of DNA in 2 specimens (patients 4 and 12). Among them, 3 patients (patients 1, 11, and 13) had received multidrug treatment for mycobacterial infections previously. Two patients (patients 1 and 13) received rifampicin, ethambutol, and clarithromycin, while another patient (patient 11) received rifampicin, levofloxacin, and clarithromycin. Patients 11 and 13 were treated in 2007 and 2013, respectively. Patient 1 had been treated since April 2018. The most recent NTM isolates for the 3 patients (patients 1, 11, and 13) prior to the M. lentiflavum isolates were identified in May 2020, December 2016, and February 2011, respectively.
Table 2. Results of the Genetic Analysis of Mycobacterium lentiflavum Isolates Identified by MALDI-TOF/MS

Note. MALDI-TOF/MS matrix-assisted laser desorption/ionization time-of-flight mass spectroscopy; NTM, nontuberculous mycobacteria; BALF, bronchoalveolar lavage fluid; Tb, tuberculosis; ILD, interstitial lung disease; SNP, single-nucleotide polymorphism.
The details of isolates including the environmental sample (patient 18) and their genetic analysis. All samples were collected in the respiratory department or the emergency department.
All isolates except no. 1 and no. 11 had genetic similarity with the environmental sample. The genetic character of isolate no. 1 was relatively close to ATCC51985 and different from no. 11.
The SNP difference between isolate nos. 1 and 11 was 35,663 and the difference between no. 1 and ATCC51985 was 62.
a Type of samples.
b Clinical diagnosis.
c Previous treatment for nontuberculous mycobacteria.
d Number of quantitative genetic difference from the SNP analysis. Lower numbers indicate a genetic similarity.
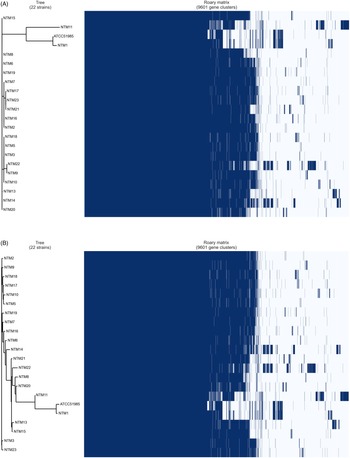
The results of the core genome and accessory analyses of the samples demonstrated polymorphism similarity, except for 2 NTM isolates (NTM1 and NTM11) from the specimens of patients 1 and 11, respectively (Table 2 and Fig. 1). The MLST analysis also showed that the 2 isolates (NTM1 and NTM11, ST646-like) were not closely related to the others (ST646). These 2 patients had a history of NTM treatment. However, no other clinical commonalities were found. A strain derived from an environmental culture (NTM18) belonged to a cluster identical with those from the clinical specimens (NTM5 and NTM10) in the core genome phylogeny (Fig. 1A). The accessory genome analysis also showed a similar observation, demonstrating that NTM18 was in the same cluster with NTM5 and NTM10 (Fig. 1B).

Fig. 1. Phylogenetic analysis based on core and accessory genomes of Mycobacterium lentiflavum isolates derived from patient and environmental samples. Core genome (A) and accessory genome (B) analyses. Right side of matrix indicated the present (dark blue) and absent (white) genes in each isolate among 9,601 gene clusters. This matrix was created by gene annotation by Prokka, and the comparison was performed by BLASTP with a percentage sequence identity (95%) which are calculated during Roary algorism. aST, sequential type. bWhole-genome frequency plot, which suggests a virtual genomic difference of each sample.
Prevention of further M. lentiflavum cases
Mycobacterium lentiflavum began to be isolated at our hospital in 2018 (Fig. 2). The period with the highest frequency of isolates was from January 2021 to March 2021, with 9 cases detected. In April 2021, we stopped using the tap that was identified to have positive environmental cultures. Distilled water was used for bronchoscope leak testing, and the part of the nebulizer that comes in contact with the patient was changed to a disposable unit. Since these interventions, the number of M. lentiflavum isolates has considerably decreased. The numbers of isolates of other NTM species have not decreased sharply in parallel with M. lentiflavum because few other species were identiefied (Supplementary Appendix Figure S1 online).

Fig. 2. Transition of the frequency of Mycobacterium lentiflavum–positive isolates.
The frequencies of M. lentiflavum–positive isolates every 4 months are shown. M. lentiflavum has been isolated from May 2018. The frequency increased gradually until April 2020. We performed the environmental survey in February 2021, and 1 sample from tap water was isolated for M. lentiflavum. After the environmental intervention, the frequency decreased considerably. The stripe bar indicates the number of the isolates after the intervention.
Discussion
As a result of the environmental survey, one sample from tap water in the respiratory medicine department was positive for M. lentiflavum. We analyzed the samples from patients and environment by WGS analysis, and we identified that most of the patient samples had a genetic similarity with the environmental sample. Because the patient isolates were deemed contaminants, we judged this episode as a pseudo-outbreak.
Among the 22 patient samples, most samples had a high genetic similarity to environmental cultures, with the exception of 2 isolates. The core-genome and accessory-genome analyses of the 2 isolates revealed differences in polymorphism, but most of the sequence types were the same, except for NTM1 and NTM11. Moreover, although the place of residence of all patients was also plotted, some lived >100 km from the hospital. Thus, we considered that the detected M. lentiflavum may have originated from the water system supplying the respiratory medicine outpatient department where the environmental culture grew the genetically related isolate.
Mycobacterium lentiflavum is a low-pathogenic bacterium.Reference Tsitko, Rahkila and Priha3–Reference Yagi, Morimoto and Ishii6 We considered it a contaminant in all but 1 case (patient 10). That patient’s isolate was treated, but her condition did not worsen although treatment was discontinued early, suggesting that her case, too, may have represented contamination. No other patient with M. lentiflavum isolates developed disease suggesting invasive infection.
Sputum and BALF were both contaminated due to use of tap water. In the environmental survey, we found that tap water passed through a bronchoscope before every examination. Because this inappropriate routine was only performed on outpatient exams, no M. lentiflavum–positive samples were collected from inpatients. Tap water was used to rinse equipment involved in collecting outpatient sputum samples. Inadequate infection control also contributed to the high number of M. lentiflavum–positive cases. Based on these results, we concluded that our hospital experienced a clinical and genetic pseudo-outbreak of at least 18 of 20 specimens. We prohibited the use of tap water from the hospital’s potable water supply system, which considerably reduced the frequency of M. lentiflavum–positive isolates. Fortunately, no patients developed significant health problems in this incident from M. lentiflavum. However, if the contaminated strain had been a more virulent species than M. lentiflavum, there could had been more serious consequences.
Restriction fragment-length polymorphism and variable-number tandem repeat methods have been used as typing methods for mycobacterial infections, but WGS analysis is becoming a standard analysis method. To the best of our knowledge, this study is the first to use the WGS method for NTM pseudo-outbreaks. In this study, the pseudo-outbreak was proven through core genome and accessory genome analyses and MLST with WGS analysis. The WGS analysis enables a systematic comparison of isolates and the detection of antimicrobial resistance gene mutations.Reference Mutayoba, Michael and Heinrich17,Reference Shanmugam, Kumar and Sembulingam18
Additionally, WGS analysis gives a more accurate judgment compared with MLST analysis alone in this study. MLST analysis uses housekeeping genes, which is independent of the specimen’s environment and refers to the specific library of a sequence type. Due to this inspection characteristic, specimens detected in an environmentally narrow range, as in this study, are more likely to be the same allele in the MLST analysis.Reference Wei, Bhushan and Stephen19 The core-genome and accessory-genome analyses compare a broad range of genes; thus, WGS analysis can be used to assess the similarity of nosocomial isolates with greater resolution. WGS analysis is a powerful method for determining the presence or absence of outbreaks.
The present study had several limitations. Prohibiting the use of tap water, which caused the pseudo-outbreak, did not eliminate the M. lentiflavum isolates completely. The fact that M. lentiflavum has also been isolated from patients whose samples were collected in an emergency room that is far from the causative water supply, suggesting that the pseudo-outbreak may not have been caused solely by the identified outpatient water supply. If environmental cultures from the other areas of the hospital test positive, it is possible that the nonhospital water-supply system is contaminated; thus, a more extensive investigation will be needed. It is also possible that, for example, the number of bacteria separated from the water supply by the creation of biofilm has increased significantly and that there is a slight amount of bacterial contamination in the entire water system used throughout the hospital. It is difficult to dismantle this water system due to its structure and cost, and it is not possible to investigate the formation of biofilm. An Australian study reported a widespread M. lentiflavum contamination of drinking water.Reference Marshall, Carter and Torbey4
Genetic analysis showed less genetic similarity in 2 samples. Although the 2 patients had in common a history of treatment for other NTM, a third patient with a history of treatment for NTM had a clonal isolate. In the genetic analysis, it is difficult to link the two isolates to the pseudo-outbreak in this study. As mentioned earlier, there may be other undiscovered sources of M. lentiflavum attributed to these samples.
In conclusion, we used the WGS analysis to genetically demonstrate a pseudo-outbreak of M. lentiflavum in our hospital. By examining environmental cultures, we found that tap water used for respiratory testing, including bronchoscopy, was the cause of this pseudo-outbreak. WGS method is more powerful than older methods to investigate pseudo-outbreaks.
Supplementary material
To view supplementary material for this article, please visit https://doi.org/10.1017/ice.2023.68
Acknowledgments
We thank Enago (https://www.enago.jp) for editing a draft of this manuscript.
Financial support
This work was supported by JSPS KAKENHI (grant no. JP20K08570).
Conflicts of interest
All authors report no conflicts of interest relevant to this article.