Introduction
Rationale
Bovine respiratory disease complex (BRD) is the most economically significant disease of feedlot cattle (Apley, Reference Apley2006; Miles, Reference Miles2009; Taylor et al., Reference Taylor, Fulton, Lehenbauer, Step and Confer2010). Putative causal organisms include Mannheimia haemolytica, Pasteurella multocida, Histophilus somni, Mycoplasma bovis, bovine herpes virus, bovine viral diarrhea virus, bovine respiratory syncytial virus, and parainfluenza type 3 virus. Although vaccination against the putative causal organisms is frequently used to help control BRD, antibiotics are also given to beef cattle when the cattle first arrive at feedlots (United States Department of Agriculture Animal and Plant Health Inspection Service Veterinary Services Centers for Epidemiology and Animal Health April, 2013). With growing concern about antibiotic use in the beef industry, it is essential to fully understand the efficacy of antibiotics to ensure that they are being used prudently (Landers et al., Reference Landers, Cohen, Wittum and Larson2012; Martin et al., Reference Martin, Thottathil and Newman2015; White House Forum Spotlights Antibiotic Overuse in Medicine and Food Production, 2015).
Drug efficacy is one of the many factors that must be considered when selecting antibiotics for use in a given setting. Evidence of the comparative efficacies of the available antibiotics is essential for prudent selection. If veterinarians know that multiple antibiotic regimes have comparable efficacies, then factors such as cost and antibiotic stewardship considerations can be used to differentiate among comparable products.
Systematic review and pairwise meta-analysis of randomized controlled trials yield the best evidence of the comparative efficacies of different treatments under field conditions (Wellman and O'Connor, Reference Wellman and O'Connor2007; Baptiste and Kyvsgaard, Reference Baptiste and Kyvsgaard2017). It is often the case, however, that an insufficient number of trials are available for a pairwise meta-analysis. In such situations, network meta-analysis can be used to borrow information from the network of available evidence in order to obtain estimates of comparative efficacy (Taylor et al., Reference Taylor, Fulton, Lehenbauer, Step and Confer2010; Jackson et al., Reference Jackson, White, Price, Copas and Riley2017; Lin et al., Reference Lin, Xing, Kofler and Murad2019). Several network meta-analyses have been conducted to evaluate the comparative efficacies of antibiotics for the treatment and control of BRD in beef production (O'Connor et al., Reference O'Connor, Coetzee, da Silva and Wang2013, Reference O'Connor, Yuan, Cullen, Coetzee, da Silva and Wang2016; Abell et al., Reference Abell, Theurer, Larson, White and Apley2017). Although the assessment of bias is an important component of a systematic review, an approach to describe the contributions of multiple studies to biases in estimates resulting from systematic reviews was not available until recently (CINeMA: Confidence in Network Meta-Analysis [Software] 2017).
Objectives
We sought to comprehensively assess the evidence for the efficacies of the different antibiotics used to control BRD in feedlot cattle in the USA using the network of trials currently available in the literature. In addition, we sought to update the approach used to assess the risk of bias in our estimates of efficacy.
Methods
Protocol and registration
The protocol for this study was developed before the review, approved by the funding agency advisory board and the funding agency staff, and posted online for public access (https://works.bepress.com/annette_oconnor/82/ and http://www.syreaf.org). Some minor modifications of the approved protocol were made to facilitate a more comprehensive systematic review and are described as needed. This review was reported using the approach described previously for network meta-analyses (Hutton et al., Reference Hutton, Salanti, Caldwell, Chaimani, Schmid, Cameron, Ioannidis, Straus, Thorlund, Jansen, Mulrow, Catala-Lopez, Gotzsche, Dickersin, Boutron, Altman and Moher2015).
Eligibility criteria
Population
The eligible population was defined as weaned cattle raised for meat production in intensive systems that put animals at risk of BRD. Eligible cattle were housed in feedlot settings, that is, as groups of penned cattle receiving rations rather than grazing on pasture. Calves explicitly described as veal or dairy calves were excluded from consideration. Calves raised on pasture, such as pre-conditioning and stocker calves, also were excluded.
Interventions
The interventions of interest were the injectable antibiotics listed in Table 1. The interventions were used in the early stages of the feedlot period to control the incidence of BRD. Any injectable antibiotic regime was eligible for inclusion; however, for reporting purposes, we only report on the efficacy of regimes that are licensed for use in the USA. All of the included antibiotics are licensed by the United States Food and Drug Administration (FDA) for the purpose of disease control, so the purpose of each antibiotic use was defined as control rather than treatment or prevention. The animals that received the antibiotics were a mixture of animals that were clinically sick but did not display detectable symptoms of BRD and animals that were disease free but at risk of developing BRD. Studies that assessed the efficacy of antibiotics used to treat cattle that had been previously diagnosed with BRD were not relevant to the review. Some studies explicitly removed detectably ‘sick’ animals from the study population at the time the animals arrived at the feedlot. The FDA considers such a study design to be an assessment of BRD control rather than BRD prevention, given the imperfect sensitivity of the methods used to detect BRD when the cattle arrive at the feedlot (Timsit et al., Reference Timsit, Dendukuri, Schiller and Buczinski2016). Although the FDA uses the term ‘control,’ we were unable to find an explicit definition of disease control used by the FDA. The American Veterinary Medical Association (AVMA) defines control as follows: Antimicrobial control of disease (synonym: metaphylaxis):
• Control is the administration of an antimicrobial to an individual animal with a subclinical infection to reduce the risk of the infection becoming clinically apparent, spreading to other tissues or organs, or being transmitted to other individuals.
• On a population basis, control is the use of antimicrobials to reduce the incidence of infectious disease in a group of animals that already has some individuals with evidence of infectious disease or evidence of infection (American Veterinary Medical Association, 2017).
Table 1. Antibiotic regimens extracted from studies identified by the systematic review
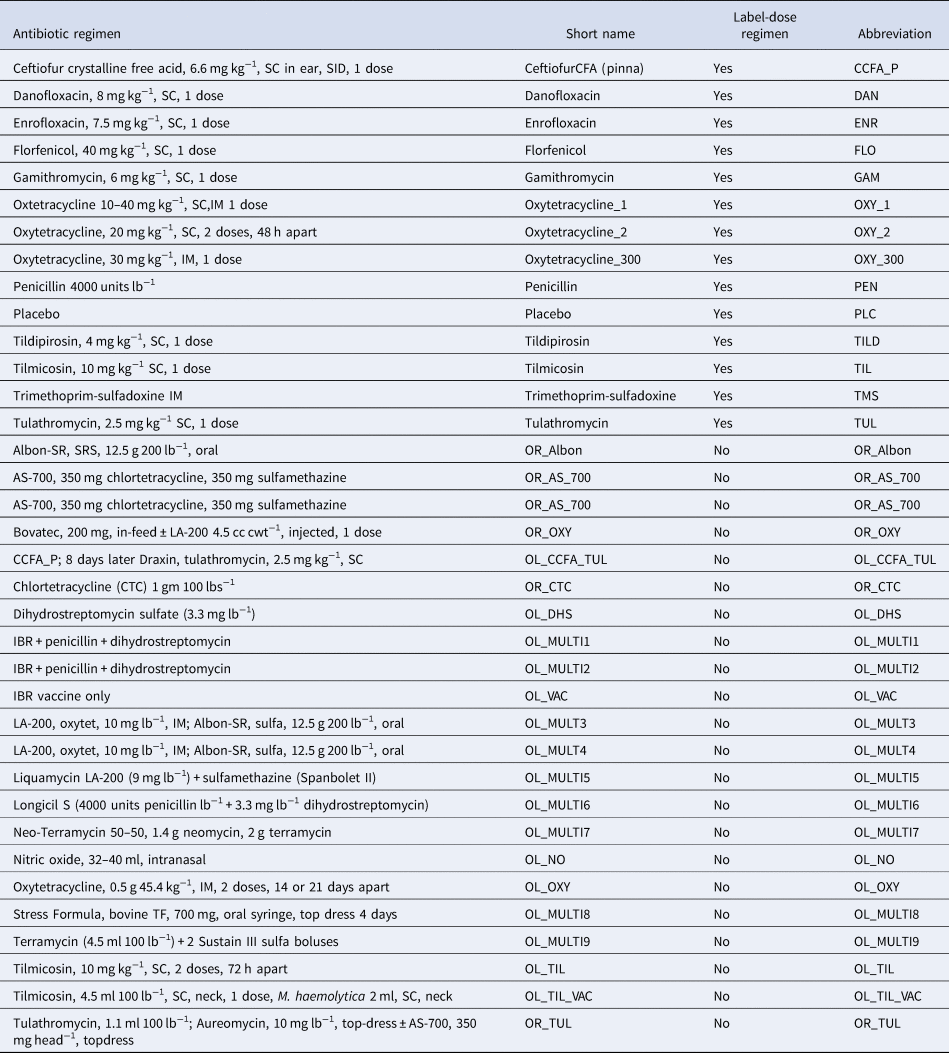
The antibiotic use in the studies included in our review corresponds to the second item in the AVMA definition. Furthermore, the synonym given by the AVMA for antimicrobial disease control, metaphylaxis, is defined by the European Union Commission as follows: term ‘metaphylaxis’, refers to the administration of the product at the same time to a group of clinically healthy (but presumably infected) in-contact animals, to prevent them from developing clinical signs, and to prevent further spread of the disease (European Union Commission, 2015), which is consistent with the AVMA definition. Much of the debate about what is control or prevention in the USA and Europe occurred after many of the studies in our review were conducted and reported. Therefore, the authors of the studies might have been uncertain about the distinctions between prevention and control, which are now important considerations but were not always so, and the peer-review process might likewise have been relatively unconcerned about ensuring that the precise purpose of the antibiotic use (i.e. prevention or control) was clarified.
The outcome that we extracted from the studies was the cumulative incidence of initial treatment for diagnosed BRD within the first 45 days that the cattle were on the feedlot. The choice of 45 days was somewhat arbitrary compared to other possible intervals such as 30 or 60 days. However, the goal was to capture early occurrences of BRD that might be impacted by the use of antibiotics at the time the cattle arrived on the feedlot. Secondary outcomes were BRD incidence and BRD mortality over the entire time the cattle were on the feedlot. The metrics extracted from the studies were prioritized as follows:
• First priority metric: Estimates of efficacy that adjusted for the clustering of feedlot populations, such as adjusted risk ratios, adjusted odds ratios, or the arm-level probability of an event obtained by transformation of the adjusted odds ratio. If the study was conducted in only one pen, the adjustment was not considered necessary.
• Second priority metric: Estimates of efficacy that did not adjust for the clustering of feedlot populations, such as unadjusted risk ratios, unadjusted odds ratios, or the arm-level probability of an event obtained by transformation of the unadjusted odds ratio.
• Third priority metric: Raw arm-level data, such as the number of animals with BRD and the number of animals allocated and analyzed in the group.
If the first priority metric was reported, the lower priority metrics were not extracted. Our rationale for the prioritization was that the meta-analysis should use an adjusted summary effect, as most relevant studies are randomized trials conducted in clustered populations.
Study designs
Studies relevant to the review had to contain at least one comparison group (active comparator or placebo) and at least one group that received one of the injectable antibiotic regimens considered an on-label use to control BRD in feedlot cattle (Table 1). Each study had to describe how the investigator selected the allocation to distinguish it from a cohort study. Challenge studies were not eligible. Although the approved protocol stated that only randomized studies were to be included, studies that reported using ‘systematic randomization’ or pure systematic (e.g. alternation) allocation were included, because they were considered trials. The potential for bias associated with non-random allocation methods was captured by the risk-of-bias assessment. Studies with cluster-based or individual-based allocation were eligible for inclusion.
Report characteristics
In addition to the eligibility criteria described in the PICOS elements described above, the included studies had to have their full texts available in English. Peer review was not an eligibility criterion, as information from conference proceedings was eligible for inclusion. No country or date restrictions were applied.
Information sources
The electronic databases used for the literature search were MEDLINE, MEDLINE In-Process and MEDLINE Daily, Epub Ahead of Print, Cambridge Agricultural and Biological Index (CABI), Science Citation Index, Conference Proceedings Citation Index Science, and AGRICOLA. MEDLINE sources were searched using the Ovid interface. AGRICOLA was searched via Proquest. The remainder of the databases were searched using the Iowa State University Web of Science interface.
The proceedings of the American Association of Bovine Practitioners (1997–2017) and the World Buiatrics Conference (1997–2017) were searched for relevant citations. We also searched the FDA Freedom of Information (FOI) New Animal Drug Applications (NADA) summaries online (https://animaldrugsatfda.fda.gov/adafda/views/#/foiDrugSummaries). European Medicines Authority (EMA) data were not searched because neither the European Public Assessment Report (EPAR) nor its product information provides data similar to the FDA FOI NADA summaries. Finally, we searched the bibliographies of the reviews that were identified in the initial screening for additional studies (Abell et al., Reference Abell, Theurer, Larson, White and Apley2017; Baptiste and Kyvsgaard, Reference Baptiste and Kyvsgaard2017). The search results were uploaded to EndNoteX9 (Clarivate Analytics, Philadelphia, PA, USA), and duplicate results were identified and removed. The records were then uploaded to the systematic review management software DistillerSR (Evidence Partners Inc., Ottawa, ON, USA) and examined further to exclude duplicate records.
Search
The searches conducted are summarized in Supplementary Tables S1–S5. The search approach was modified for the different database or information source as appropriate.
Selection of studies relevant to the review
The first round of study selection was based on abstracts and titles. The second round of study selection, for relevance to the meta-analysis, was based on the full text of the articles. The questions used for each level of screening are provided in the protocol available online (https://works.bepress.com/annette_{o}connor/82/). Some minor modifications for clarity were made to the questions in the protocol, as described in the text accompanying the Supplementary materials S1 and S12. For example, an additional answer option was included that allowed reviewers to identify reviews that were potentially relevant to the project.
Two reviewers conducted a pre-test of the first 100 abstracts identified in the search to ensure that the questions were clear and consistently understood. The questions and their wording were revised during the pre-test phase until there was agreement that they captured the required information. Studies were excluded if both reviewers responded ‘no’ to either Q1 or Q2 in Table S2. If at least one reviewer responded ‘yes’ to each question, the study was advanced to the full-text assessment, which determined if the studies met the eligibility criteria for inclusion in the meta-analysis. Two reviewers conducted a pre-test on the first five full texts that were advanced to the full-text assessment to again ensure that the questions were clear and consistently understood. The questions and their wording were revised further during the second pre-test phase until there was agreement that they captured the required information. During the data extraction phase, if disagreements about eligibility occurred, a third reviewer (AOC) was consulted. As a result of the consultation process, the final agreement among the reviewers was 100%.
Studies that passed the full-text assessment were included in the meta-analysis if they contained sufficient data for the log odds ratio and the standard error of the log odds ratio to be calculated using the prioritized metrics extracted from the study. The criteria for inclusion in the meta-analysis are described further in the statistical analysis section.
Data collection process
Systematic review management software (DistillerSR®, Evidence Partners, Ontario, Canada) was used to extract data from the selected studies into the pre-tested forms. Two reviewers independently extracted all of the data elements of interest from the relevant full-text articles. After extraction, any disagreements were resolved by discussion. If the discussions did not lead to consensus, a third reviewer (AOC) was consulted. The unit of concern for data extraction was the study level, if available. Data from multi-site studies were extracted at the site level if the relevant information was reported. If investigators combined multiple sites into a single analysis and only reported the pooled information, then the pooled information was extracted. We did not contact the study authors about missing data. If multiple studies were linked, we used all of the available information and cited the study that provided the most complete report.
Data items
Items such as the country and year in which the study was conducted, the interventions that were used, and the outcomes that were measured were extracted at the study level. If baseline characteristics and information about the loss to follow-up were reported, those were extracted as arm-level data. Other data extracted at the arm level included the treatments and the results (based on prioritization). The published protocol, which includes a list of all items extracted, is available online.
Geometry of the network
Network geometry was assessed using a previously proposed approach (Salanti et al., Reference Salanti, Kavvoura and Ioannidis2008). The probability of an inter-species encounter (PIE) index was calculated using a custom R script. The PIE index is a continuous variable that decreases in value as unevenness increases. Values <0.75 can be considered to reflect a limited diversity of interventions. We also assessed co-occurrence using the C-score, which is based on a checkerboard analysis and describes whether particular pairwise comparisons of specific treatments are preferred or avoided within the network (Salanti et al., Reference Salanti, Kavvoura and Ioannidis2008). The C-score test was performed using the R package EcoSimR version 0.1.0 (Gotelli and Entsminger, Reference Gotelli and Entsminger2001).
Risk of bias within individual studies
The risk-of-bias form used for this study was based on the Cochrane Risk of Bias 2.0 tool for randomized trials. The form was modified to ensure its relevance to the topic area (Higgins et al., Reference Higgins, Sterne, Savović, Page, Hróbjartsson, Boutron, Reeves and Eldridge2016). The risk-of-bias assessment was conducted at the outcome level (BRD morbidity and mortality) for the outcome assessment domain. For the other risk domains, the responses for BRD morbidity and mortality were considered the same within a given study. For the purpose of assessing the risk of bias due to allocation processes, the Cochrane Risk of Bias tool for individually allocated studies places substantial emphasis on allocation concealment. It is unclear, however, if that emphasis is applicable in beef production settings. Therefore, rather than make an overall bias assessment for bias arising from the allocation approach, we asked the following three signaling questions:
• SQ 1.1 – Was the allocation sequence random?
• SQ 1.2 – Was the allocation sequence concealed until participants were recruited and assigned to interventions?
• SQ 1.3 – Were there evident baseline imbalances that suggested a problem with the randomization process?
For cluster-randomized trials, the first three signaling questions for bias arising from the allocation approach were also assessed and presented with the individual-level questions. Two additional questions related to bias arising from the characteristics of individual participants in the cluster-randomized studies were assessed and presented separately.
• Were all the characteristics of the individual participants likely to be evenly distributed across the treatment groups?
• Were there any baseline imbalances that suggested differential identification or recruitment of individual participants between arms?
If studies within a single citation had different characteristics that impacted bias, such as loss to follow-up, a risk-of-bias assessment was made for each study separately. Otherwise, multiple studies presented within a single citation were considered as a single set of results.
The other bias domains considered were the following:
(1) Domain 2: Bias arising from deviations from the intended interventions;
(2) Domain 3: Bias arising from missing outcome data;
(3) Domain 4: Bias arising from the measurement of the outcome; and
(4) Domain 5: Bias arising from the selection of the reported outcome.
Summary measures
The baseline risk used to convert the odds ratios to the risk ratio was obtained using the distribution of the log odds of the placebo group. The posterior distribution of the mean was N(−0.3759; 1.0323). The posterior distribution of the standard deviation was N(1.0059; 0.1392).
Planned method of statistical analysis
The proposed method of statistical analysis has been described in detail elsewhere (Dias et al., Reference Dias, Welton, Caldwell and Ades2010). We used a random-effects Bayesian model for continuous outcomes. Let b denote the baseline treatment of the whole network (usually placebo), and let b i denote the trial-specific baseline treatment of trial i. It could be the case that b ≠ b i. Suppose there are L treatments in a network. Assume a normal distribution for the continuous measure of the treatment effects of arm k relative to the trial-specific baseline arm b i in trial i, $y_{ib_ik}$ , with variance
, with variance  $V_{ib_ik}$, such that
$V_{ib_ik}$, such that
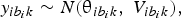 $$y_{ib_ik}\sim N\lpar {{\rm \theta }_{ib_ik}\comma \;V_{ib_ik}} \rpar \comma \;$$
$$y_{ib_ik}\sim N\lpar {{\rm \theta }_{ib_ik}\comma \;V_{ib_ik}} \rpar \comma \;$$and
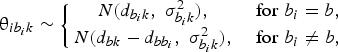 $${\rm \theta }_{ib_ik}\sim \left\{{\matrix{ {N\lpar {d_{b_ik}\comma \;\sigma_{b_ik}^2 } \rpar \comma \;} & {{\rm for\;}b_i = b\comma \;} \cr {N\lpar {d_{bk}-d_{bb_i}\comma \;\sigma_{b_ik}^2 } \rpar \comma \;} & {{\rm for\;}b_i\ne b\comma \;} \cr } } \right.$$
$${\rm \theta }_{ib_ik}\sim \left\{{\matrix{ {N\lpar {d_{b_ik}\comma \;\sigma_{b_ik}^2 } \rpar \comma \;} & {{\rm for\;}b_i = b\comma \;} \cr {N\lpar {d_{bk}-d_{bb_i}\comma \;\sigma_{b_ik}^2 } \rpar \comma \;} & {{\rm for\;}b_i\ne b\comma \;} \cr } } \right.$$
where d bk is the treatment effects of k relative to the network baseline treatment b and  ${\rm \sigma }_{b_ik}^2 $ is the between-trial variance. The priors of d bk and
${\rm \sigma }_{b_ik}^2 $ is the between-trial variance. The priors of d bk and  ${\rm \sigma }_{b_ik}$ are
${\rm \sigma }_{b_ik}$ are
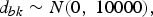 $$d_{bk}\sim N\lpar {0\comma \;10000} \rpar \comma \;$$
$$d_{bk}\sim N\lpar {0\comma \;10000} \rpar \comma \;$$
and there is a homogeneous variance assumption that  ${\rm \sigma }_{b_ik}^2 = {\rm \sigma }^2$, where σ ~ U(0, 5). Thus, for L treatments, we have L − 1 priors for d bl, l ∈ {1, …, L}, l ≠ b. For l = b, we have d bb = 0.
${\rm \sigma }_{b_ik}^2 = {\rm \sigma }^2$, where σ ~ U(0, 5). Thus, for L treatments, we have L − 1 priors for d bl, l ∈ {1, …, L}, l ≠ b. For l = b, we have d bb = 0.
Handling of multi-arm trials
For multi-arm trials, we assumed that the co-variance between  ${\rm \theta }_{jb_jk}$ and
${\rm \theta }_{jb_jk}$ and  ${\rm \theta }_{jb_j{k}^{\prime}}$ was σ2/2 (Higgins and Whitehead, Reference Higgins and Whitehead1996; Lu and Ades, Reference Lu and Ades2004). The likelihood of a trial i with a i arms would be defined as multivariate normal:
${\rm \theta }_{jb_j{k}^{\prime}}$ was σ2/2 (Higgins and Whitehead, Reference Higgins and Whitehead1996; Lu and Ades, Reference Lu and Ades2004). The likelihood of a trial i with a i arms would be defined as multivariate normal:
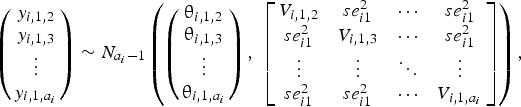 $$\left({\matrix{ {y_{i\comma 1\comma 2}} \cr {y_{i\comma 1\comma 3}} \cr \vdots \cr {y_{i\comma 1\comma a_i}} \cr } } \right)\sim N_{a_i-1}\left({\left({\matrix{ {{\rm \theta }_{i\comma 1\comma 2}} \cr {{\rm \theta }_{i\comma 1\comma 3}} \cr \vdots \cr {{\rm \theta }_{i\comma 1\comma a_i}} \cr } } \right)\comma \;\left[{\matrix{ {V_{i\comma 1\comma 2}} & {se_{i1}^2 } & \cdots & {se_{i1}^2 } \cr {se_{i1}^2 } & {V_{i\comma 1\comma 3}} & \cdots & {se_{i1}^2 } \cr \vdots & \vdots & \ddots & \vdots \cr {se_{i1}^2 } & {se_{i1}^2 } & \cdots & {V_{i\comma 1\comma a_i}} \cr } } \right]} \right)\comma$$
$$\left({\matrix{ {y_{i\comma 1\comma 2}} \cr {y_{i\comma 1\comma 3}} \cr \vdots \cr {y_{i\comma 1\comma a_i}} \cr } } \right)\sim N_{a_i-1}\left({\left({\matrix{ {{\rm \theta }_{i\comma 1\comma 2}} \cr {{\rm \theta }_{i\comma 1\comma 3}} \cr \vdots \cr {{\rm \theta }_{i\comma 1\comma a_i}} \cr } } \right)\comma \;\left[{\matrix{ {V_{i\comma 1\comma 2}} & {se_{i1}^2 } & \cdots & {se_{i1}^2 } \cr {se_{i1}^2 } & {V_{i\comma 1\comma 3}} & \cdots & {se_{i1}^2 } \cr \vdots & \vdots & \ddots & \vdots \cr {se_{i1}^2 } & {se_{i1}^2 } & \cdots & {V_{i\comma 1\comma a_i}} \cr } } \right]} \right)\comma$$
where the diagonal elements in the variance–covariance matrix represent the variances of the treatment differences, and the off-diagonal elements represent the observed variance in the control arm in trial i, denoted by  $se_{i1}^2 $. For all studies, the results were converted to log odds ratios for analysis. If the study authors reported a risk ratio, that was converted back to the log odds ratio using the reported risk of disease in the placebo group. When the authors reported the probability of BRD in each treatment arm on the basis of a model, then that probability was converted back to the logs odds ratio using a method described elsewhere (Hu et al., Reference Hu, Wang and O'Connor2019).
$se_{i1}^2 $. For all studies, the results were converted to log odds ratios for analysis. If the study authors reported a risk ratio, that was converted back to the log odds ratio using the reported risk of disease in the placebo group. When the authors reported the probability of BRD in each treatment arm on the basis of a model, then that probability was converted back to the logs odds ratio using a method described elsewhere (Hu et al., Reference Hu, Wang and O'Connor2019).
Selection of prior distributions in the Bayesian analysis
The prior distributions were originally based on an approach reported previously (Dias et al., Reference Dias, Welton, Sutton and Ades2011). As in previous models, we assessed σ ~ U(0, 2) and σ ~ U(0, 5), and our results suggested that σ ~ U(0, 5) was preferable. We repeated that assessment and retained the same prior distributions used in the previous models (O'Connor et al., Reference O'Connor, Yuan, Cullen, Coetzee, da Silva and Wang2016).
Implementation and output
All posterior samples were generated using a Markov Chain Monte Carlo (MCMC) simulation implemented with the Just Another Gibbs Sampler (JAGS) software (Plummer, Reference Plummer2015). All statistical analyses were performed using R software (version 3.5.2) (R Core Team, 2015). We fitted the model using JAGS, an MCMC sampler, by calling JAGS from R through the rjags package (version 4-8) (Plummer, Reference Plummer2015). Three chains were simulated and the convergence was assessed using Gelman–Rubin diagnostics. We discarded 5000 ‘burn-in’ iterations based on our inferences from a further 10,000 iterations. The model output included all possible pairwise comparisons of the log odds ratios (for inconsistency assessment), risk ratios (used for comparative efficacy reporting), and treatment failure rankings (for comparative efficacy reporting).
Assessment of the model fit
The fit of the model was assessed on the basis on the log odds ratios by examining the residual deviance between the predicted values from the MTC model and the observed values from each study (Dias et al., Reference Dias, Welton, Caldwell and Ades2010).
Assessment of inconsistency
Network meta-analysis relies on an assumption of consistency between direct and indirect intervention effects that are distinct from the usual variation that stems from a random effects meta-analysis model. For example, if one study compares the direct effect of treatment A with the effect of treatment B, and another study compares the efficacies of treatments B and C, then the (indirect) effect of treatment A relative to the efficacy of treatment C can be inferred. We used the back calculation method to assess the consistency assumption (Dias et al., Reference Dias, Welton, Caldwell and Ades2010). We did not rely only on the P-values for the consistency evaluation; instead, we compared the direction and magnitude of the estimates. We also compared the estimates from the direct and indirect models and considered the standard deviation of each estimate. Comparisons in which the direct and indirect estimates had different signs were further evaluated and discussed.
Risk of bias assessment across studies
To describe the overall quality of the evidence network and the bias across the studies, we employed a modification of the GRADE approach for network meta-analysis (Salanti et al., Reference Salanti, Del Giovane, Chaimani, Caldwell and Higgins2014; Papakonstantinou et al., Reference Papakonstantinou, Nikolakopoulou, Rucker, Chaimani, Schwarzer, Egger and Salanti2018) using the Confidence in Network Meta-Analysis (CINEMA) online software (http://cinema.ispm.ch) (CINeMA: Confidence in Network Meta-Analysis [Software] 2017). CINEMA takes a frequentist approach to the calculation of treatment effects based on the metafor package in R (Viechtbauer, Reference Viechtbauer2010). The contribution matrix for the risk of bias is based on that approach. An equivalent tool using a Bayesian analysis is not currently available. The system that we used evaluates within-study bias, across-studies bias, indirectness, imprecision, heterogeneity, and incoherence. We evaluated and reported the contributions of studies to the within-study bias on the basis of randomization and blinding rather than an overall assessment of bias. The rationale for using that approach was that there is evidence from veterinary science that failure to include randomization and blinding elements in the study design is associated with larger estimates of effects, whereas there is no evidence that other elements included in the Cochrane risk of bias, such as allocation concealment, introduce any bias in livestock production data (Sargeant et al., Reference Sargeant, Elgie, Valcour, Saint-Onge, Thompson, Marcynuk and Snedeker2009). For randomization, we evaluated the risk of bias as follows:
• ‘Low risk of bias’ refers to studies that reported and provided evidence of random allocation, identified by a ‘yes’ or ‘probably yes’ answer to risk-of-bias question Q1.1.
• ‘Unclear risk of bias’ refers to studies that reported random allocation but provided no evidence, identified by a ‘no information random’ response to risk-of-bias question Q1.1.
• ‘High risk of bias’ refers to studies that used non-random allocation or provided no information about allocation, identified by a ‘no,’ ‘probably no,’ or ‘no information at all’ response to risk-of-bias question Q1.1.
We considered proper blinding of caregivers and outcome assessors to be associated with a low risk of bias. If the study authors mentioned only one of those, we considered it unclear whether or not there was potential for bias, and if neither was reported, we considered the potential for bias to be high.
• ‘Low risk of bias’ refers to studies that reported blinding of caregivers and outcome assessors, identified by a ‘no’ or ‘probably no’ response to risk-of-bias questions Q 2.2 and Q4.1.
• ‘Unclear risk of bias’ refers to studies that reported blinding of caregivers and outcome assessors, identified by a ‘no’ or ‘probably no’ response to risk-of-bias question Q 2.2 or Q4.1, but not both.
• ‘High risk of bias’ refers to studies that did not fall into the above two categories.
In CINEMA, indirectness refers to how closely the study populations resemble the populations in which the intervention is to be used. Given the narrow eligibility criteria for our meta-analysis, we did not consider indirectness to be an issue. Accordingly, we considered the risk of bias due to indirectness to be low in all of the selected studies. The ability to assess bias across studies in a network meta-analysis is poorly developed. None of the studies that we reviewed had a sufficient number of pairs to make an informative pairwise assessment of small-study effects, so we did not attempt that assessment. For the assessment of imprecision, which indicates if the boundaries of the confidence intervals for the treatment effects could lead to different outcomes, we used 0.8, a clinically important odds ratio that equates to ±0.2231 on the log scale. We used the same odds ratio to assess heterogeneity. We did not present the inconsistency analysis from CINEMA, because that analysis was already conducted as part of the Bayesian analysis.
Additional analyses
No additional analyses were conducted.
Results
Study selection
The flow of the studies from citation retrieval to inclusion in the meta-analysis is shown in the PRISMA diagram in Fig. 1. In the PRISMA diagram, we only show unique relevant citations. Some citations had multiple trials, multiple arms, or multiple time periods at which outcomes were reported. Because of the rarity of the mortality outcome, we deviated from the approved protocol and did not analyze that outcome. Eighty-five trials from a total of 73 citations reported at least one BRD morbidity outcome.
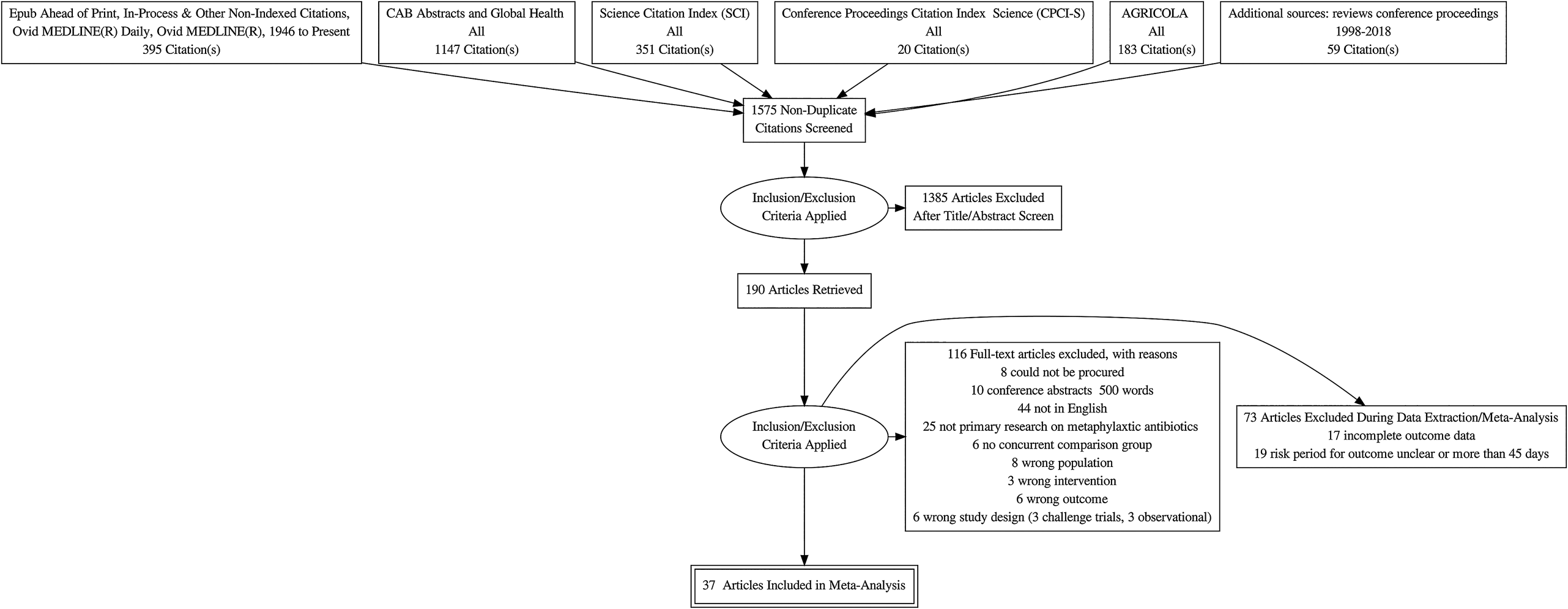
Fig. 1. Flowchart describing the flow of literature through the review.
Study selection: excluded because of data extraction issues
Seventeen citations had at least one trial in which an arm was excluded from our meta-analysis because of one of the following data issues: multiple arms represented the same treatment, there were zero events in one arm, no extractable data were presented, there was no P-value or baseline risk available for data conversion, the treatment regimen was inadequately described, or the treatment was entirely disconnected from the network (Schipper and Kelling, Reference Schipper and Kelling1974; Janzen and McManus, Reference Janzen and McManus1980; Brown et al., Reference Brown, Faulkner and Cmarik1989; van Koevering et al., Reference van Koevering, Gill, Smith, Hill and Ball1992; Guichon et al., Reference Guichon, Booker and Jim1993; Galyean et al., Reference Galyean, Gunter and Malcolm-Callis1995; Montague et al., Reference Montague, Smith and Gill1996; Reece and Smith, Reference Reece and Smith1996; Brazle, Reference Brazle1997; Hibbard et al., Reference Hibbard, Robb, Chester, Dame, Moseley and Bryson2002; Booker et al., Reference Booker, Schunicht, Guichon, Jim, Wildman, Pittman and Perrett2006; Booker et al., Reference Booker, Abutarbush, Schunicht, Jim, Perrett, Wildman, Guichon, Pittman, Jones and Pollock2007; Fazzio et al., Reference Fazzio, Giuliodori, Galvan, Streitenberger and Landoni2015; Crepieux et al., Reference Crepieux, Miller, Regev-Shoshani, Schaefer, Dorin, Alexander and Timsit2016; Miller et al., Reference Miller, Hubbert, Reinhardt, Loest, Schwandt and Thomson2016; Timsit et al., Reference Timsit, Hallewell, Booker, Tison, Amat and Alexander2017a, Reference Timsit, Workentine, Crepieux, Miller, Regev-Shoshani, Schaefer and Alexander2017b; Food and Drug Administration, 2018f).
Study selection: excluded because of the duration of BRD assessment
Nineteen trials either did not report the duration of their observations of BRD incidence or had an observation period longer than 45 days (Schipper and Kelling, Reference Schipper and Kelling1971; Stokka and Kreikemeier, Reference Stokka and Kreikemeier1996; Schunich et al., Reference Schunich, Guichon, Booker, Jim, Wildman, Hill, Ward, Bauck and Jacobsen2002; Cusack, Reference Cusack2004; Step et al., Reference Step, Engelken, Romano, Holland, Krehbiel, Johnson, Lawrence, Tucker and Robb2007; Bryant et al., Reference Bryant, Nichols, Adams, Farmer and Miles2008; John et al., Reference John, Bryson, Sam and Breck2008; van Donkersgoed et al., Reference van Donkersgoed, Merrill and Hendrick2008; Corbin et al., Reference Corbin, Gould, Carter, McClary and Portillo2009; Blasi et al., Reference Blasi, Epp and Derstein2010; van Donkersgoed and Merrill, Reference van Donkersgoed and Merrill2012; Hendrick et al., Reference Hendrick, Bateman and Rosengren2013; Stegner et al., Reference Stegner, Lucas, McLaughlin, Davis, Alaniz, Weigel, Pollreisz, Tucker, Koers, Turgeon and Szasz2013; van Donkersgoed and Merrill, Reference van Donkersgoed and Merrill2013a, Reference van Donkersgoed and Merrill2013b; Compiani et al., Reference Compiani, Baldi, Bonfanti, Fucci, Pisoni, Jottini and Torres2014; Tennant et al., Reference Tennant, Ives, Harper, Renter and Lawrence2014; Sturgess and Renter, Reference Sturgess and Renter2017; van Donkersgoed et al., Reference van Donkersgoed, Hendrick and Nickel2017).
Study selection: included in the meta-analysis
Our meta-analysis included data from 37 citations that described 46 studies in which BRD morbidity was reported within 45 days of the arrival of the cattle at the feedlot (Lofgreen et al., Reference Lofgreen, Addis, Dunbar and Clark1978; Breeze et al., Reference Breeze, McManus, Magonigle, Olson, Tonkinson and Costerton1982; Gill et al., Reference Gill, Smith, Hicks and Ball1986; Schumann et al., Reference Schumann, Janzen and McKinnon1990; Harland et al., Reference Harland, Jim, Guichon, Townsend and Janzen1991; Schumann et al., Reference Schumann, Janzen and McKinnon1991; Morck et al., Reference Morck, Merrill, Thorlakson, Olson, Tonkinson and Costerton1993; Vogel et al., Reference Vogel, Laudert, Zimmermann, Guthrie, Mechor and Moore1998; McClary and Vogel, Reference McClary and Vogel1999; Duff et al., Reference Duff, Walker, Malcolm-Callis, Wiseman and Hallford2000; Frank and Duff, Reference Frank and Duff2000; Glynn et al., Reference Glynn, Robert, Glenn, Raymond and Charles2002; Meeuwse and Hibbard, Reference Meeuwse and Hibbard2002; Food and Drug Administration, 2003; Guthrie et al., Reference Guthrie, Rogers, Christmas, Vogel, Laudert and Mechor2004; Kilgore et al., Reference Kilgore, Spensley, Sun, Nutsch, Rooney and Skogerboe2005; Rooney et al., Reference Rooney, Nutsch, Skogerboe, Weigel, Gajewski and Kilgore2005; Martin et al., Reference Martin, Partida, Villalobos, Lopez, Lopez-Guerrero and Blanco2007; Benton et al., Reference Benton, Erickson, Klopfenstein, Luebbe, Smith, Sides and Bryant2008; Montgomery et al., Reference Montgomery, Sindt, Greenquist, Loe and Drouillard2008; Nickell et al., Reference Nickell, White, Larson, Blasi and Renter2008; Rossi et al., Reference Rossi, Vandoni, Bonfanti and Forbes2010; Baggott et al., Reference Baggott, Casartelli, Fraisse, Manavella, Marteau, Rehbein, Wiedemann and Yoon2011; Gonzalez-Martin et al., Reference Gonzalez-Martin, Elvira, Lopez, Villalobos, Lopez-Guerrero and Astiz2011; Siddartha et al., Reference Siddartha, Thomson, Bello, Nosky and Reinhardt2013; Amrine et al., Reference Amrine, White, Goehl, Sweiger, Nosky and Tessman2014; White et al., Reference White, Amrine and Goehl2015; DeDonder et al., Reference DeDonder, Apley, Li, Gehring, Harhay, Lubbers, White, Capik, KuKanich, Riviere and Tessman2016; Hanzlicek et al., Reference Hanzlicek, Blasi, Oleen and Anderson2016; Hawley et al., Reference Hawley, Powell, Kegley and Beck2016; Regev-Shoshani et al., Reference Regev-Shoshani, McMullin, Nation, Church, Dorin and Miller2017; Food and Drug Administration, 2018a, 2018b, 2018c, 2018d, 2018e, 2018f, 2018g).
Presentation of the network structure
The final evidence network used in the meta-analysis represented 46 studies and 167 arms. Some of the arms used treatment regimens that were not labeled for use in the USA but contributed useful information about variations of regimens of interest. The antibiotic regimens used in those off-label studies are reported in Table 1. Four of the included studies were three-arm studies, and one was a four-arm study. Thirty-four of the studies compared non-active or controlled treatment groups with active treatment groups, whereas the other 12 studies compared multiple active treatment groups.
Summary of the network geometry
The geometry of the network was sparse, with many regimens assessed only once. The network was diverse, however, as indicated by the PIE index of 0.84. A PIE index of >0.75 suggests that a network is diverse, although, as with any threshold, that is a guideline rather than a strict cut-off for interpretation purposes (Salanti et al., Reference Salanti, Kavvoura and Ioannidis2008). The conclusion that the network was diverse is consistent with a visual examination of the network, which includes a large number of treatments (Fig. 2). The diversity measure considers all treatments included in the meta-analysis and is bolstered by treatments that were not relevant to the meta-analysis. The C-score was 19.45 (P = 0.43). The C-score is used to evaluate how random encounters occur in ecological populations. When used in a network meta-analysis, the C-score assesses whether or not there are particular pairwise comparisons that occur more (or less) often than expected if the pairing is random. Although the P-value suggests that the pairing was random, visual examination of the network suggests that researchers had a preference for comparisons that involved placebo-controlled study arms.

Fig. 2. The network of treatment arms used in the mixed-treatment comparisons meta-analysis. The size of the dot is a relative indicator of the number of arms, and the width of the lines is a relative indicator of the number of direct comparisons. Abbreviations defined in Table 1.
Study characteristics
The descriptive information, including the definitions of success and the exclusion criteria, about the studies identified in the literature search is provided in the Supplementary materials (Table S6). All of the relevant studies, rather than just those included in the meta-analysis, are listed in Table S6 to show how the study populations differed between the studies that were included in the meta-analysis and those that were excluded. Of particular interest is the baseline conditions applied to the animals in each study. Some of the studies failed to clearly document concurrent treatments that were received by the animals, such as vaccinations, antiparasitic treatments, and antibiotics. Other studies did provide that information, and some even reported using in-feed antibiotics concurrently with the injectable antibiotics for BRD control.
The approaches used to handle the cases of BRD that were diagnosed at the time the cattle arrived on the feedlot are reported in Table S7. Many studies failed to clearly describe how those cases of BRD were handled. The definitions of BRD reported by the study authors are listed in Table S8. In each instance where a single citation included multiple studies, the studies used the same definition of the outcome, so Table S8 is indexed by citation rather than by study. The definitions of outcomes were generally consistent and frequently reported.
Individual risk of bias
The results of the risk-of-bias assessments for the studies that were determined to be relevant to the meta-analysis are shown in Table S9. The answers to the two additional questions for the cluster-randomized trials are presented in Table S10.
Synthesis of the results
The final meta-analysis included results from 46 studies. The measures of convergence from the Bayesian model were within normal limits. The results of the model are presented in several ways. Table 4 shows the comparative risk ratios for only the label-dose regimens. The data are organized so that the event is the risk of treatment failure for the treatment listed in the row, divided by the risk of failure for the treatment listed in the column. For example, in the first row of the table, all the risk ratio estimates are >1, meaning that the risk of treatment failure was higher in the non-active control (NAC) groups for PLC compared with that in the active treatment groups for each of the other antibiotics. The upper right-hand quadrant of Table 4 shows the estimated risk ratio, and the lower quadrant reports the 95% CI. Estimates of the average ranks of the antibiotics in terms of efficacy are provided in Fig. 3, which only includes the label-dose regimens. The rankings for the label-dose regimens in the meta-analysis are presented in Table 2. We excluded the off-label regimens from Fig. 3 because we do not want to promote the off-label use of antibiotics. Rankings close to 1 are associated with low rates of treatment failure. There is considerable overlap in the confidence intervals of the rankings. The probability distribution of the treatment responses for the label-dose protocols is presented in the Supplementary materials (Table S13 and Fig. S1). Also included in the Supplementary materials is a summary of the probabilities of being ranked first and last (Table S14) and all possible pairwise probabilities of being better or worse than other antibiotic regimens in Table S15.

Fig. 3. Ranking plot of relevant treatments. Lower rankings indicate lower incidence of BRD. Rankings are reported for label-dose regimens only. Abbreviations are defined in Table 1.
Table 2. Mean ranking by treatment efficacy of antibiotic regimens based on the mixed-treatment comparisons meta-analysis

Lower rankings indicate a lower incidence of BRD. World Health Organization categories for medically important antibiotics: Critically Important Antimicrobial (CIA), Highly Important Antimicrobial (HIA), Important Antimicrobial (IA) (SD, standard deviation).
Exploration of inconsistency
The consistency between the direct and indirect sources of evidence in the final model using 46 studies and 167 arms is reported in Table 3. In the model, we found no evidence of inconsistency between the direct and indirect estimates. A potential reason for that finding is that the small number of studies available for some comparisons caused the standard deviations for the direct estimates to be very wide, making it difficult to detect differences between direct and indirect estimates.
Table 3. Results of the indirect comparison for the consistency assumption

Posterior means (d) and standard deviations (SD) of the log-odds ratio of treatment effects calculated using direct evidence only (dir), all evidence (MTC), and indirect evidence only (rest). The treatment on the left is the reference (denominator), and that on the right is the comparator (numerator). w and SD(w) are the inconsistency estimate and standard deviation of the inconsistency estimate, respectively.
Risk of bias across studies
There was an enormous number of possible pairwise comparisons (i.e. all possible pairs) in the risk-of-bias assessment. Therefore, we only presented the active-to-active comparisons of on-label antibiotic regimes (Table S11). The contributions of the included studies to the risk of bias based on the studies' randomization status are presented in Fig. 4. The contributions based on the studies' blinding status are presented in Fig. 5. Fewer studies presented a risk of bias due to randomization issues than presented risks due to blinding issues. We assumed that all of the studies were free of bias related to indirectness. For each outcome, a considerable number of comparisons presented major concerns related to imprecision, reflecting the large confidence intervals of the risk ratios resulting from the small number of trials in each comparison. Most of the comparisons presented no concerns about bias due to heterogeneity, which was expected on the basis of the wide confidence intervals of the risk ratios.

Fig. 4. The contributions to the point estimate of efficacy by studies using different allocation approaches: green indicates studies that provided evidence of random allocation; yellow indicates studies that reported random allocation but provided no supporting evidence; red indicates studies that reported no allocation approach or reported a non-random allocation approach. White vertical lines indicate the percentage contribution of separate studies. Each bar shows the percentage contribution from studies judged to be at low (green), moderate (yellow), and high (red) risk of bias.

Fig. 5. The contributions to the point estimate of efficacy by studies using different blinding approaches: green indicates studies that provided evidence of blinding of caregivers and outcome assessors; yellow indicates studies that provided evidence of blinding of either caregivers or outcome assessors; red indicates studies that reported no blinding of caregivers or outcome assessors. White vertical lines indicate the percentage contribution of separate studies. Each bar shows the percentage contribution from studies judged to be at low (green), moderate (yellow), and high (red) risk of bias.
Results of additional analyses
No additional analyses were conducted.
Discussion
Summary of the evidence
The results suggest that the macrolides as a group are the most effective antibiotics administered to cattle upon arrival at the feedlot to control BRD. The evidence for that conclusion is summarized best in the ranking plot (Fig. 3), which shows that the regimens involving tulathromycin, gamithromycin, tilmicosin, and tildipirosin are the top-ranked treatments. Ceftiofur had the next highest ranking, after the macrolides. Those results are consistent with the findings of other reviews of antibiotic use to control BRD in feedlot cattle (Abell et al., Reference Abell, Theurer, Larson, White and Apley2017; Baptiste and Kyvsgaard, Reference Baptiste and Kyvsgaard2017). The macrolides are listed as critically important antibiotics with the highest priority by the World Health Organization (WHO). Oxytetracycline and florfenicol, also listed as important antibiotics by the WHO, were less effective than the macrolides and ceftiofur, although they were more effective than danofloxacin and enrofloxacin (both of which are critically important). Many of the comparisons with other regimens in our meta-analysis are based on only a single study, so confidence in the pairwise differences is low. Data from the National Animal Health Monitoring Scheme Antibiotic Use 2017 report indicate that 14.8% (SE 3.5) of feedlots used injectable antibiotics in groups of cattle. In that report, ‘treated as a group’ meant that at least 90% of the cattle in a pen or group were treated with one or more injectable antimicrobial agent for the purpose of preventing, controlling, or treating a disease outbreak. Injectable oxytetracycline is the antibiotic most commonly used on groups of cattle on feedlots in the USA (United States Department of Agriculture Animal and Plant Health Inspection Service Veterinary Services National Animal Health Monitoring System May, 2019) (4.8%, SEM 2.5), followed by tulathromycin (2.8%, SEM 0.8), ceftiofur (2.7%, SEM 1.3), and enrofloxacin (2.4%, SEM 2.0). In terms of the percentage of cattle treated, rather than the percentage of feedlots using the antibiotic, the injectable antibiotic most commonly used in groups of cattle on feedlots was tulathromycin (4.4%, SEM 0.6), followed by gamithromycin (2.9%, SEM 0.8). It would be tempting to link those statistics with the estimates of efficacy from our network meta-analysis in order to make inferences about prudent antibiotic choices. That is not feasible, however, because the NAHMS report does not distinguish among instances of injectable antibiotic use for prevention, control, and treatment of BRD. Our results suggest that most of the injectable antibiotics are very effective at reducing the incidence of BRD during the first 45 days that cattle are on the feedlot. Although the posterior distributions of the risk ratios were often wide, as suggested by the credible intervals in Table 4, the means of the posterior distribution of the risk distribution suggest that apart from sulfamethoxazole and trimethoprim combinations, the injectable antibiotics reduced the risk of BRD within the first 45 days on the feedlot by 1.5-fold on average compared with placebo. An alternative way to look at those data is to use Table S15, which shows that all of the antibiotic regimens have at least a 0.5 probability of being better than a NAC. Because the diagnosis of BRD is imperfect, and misclassification would bias the associations toward the null, the actual efficacy of the antibiotics is likely even higher than our estimates (White and Renter, Reference White and Renter2009).
Table 4. Risk ratio of all possible pairwise comparisons within the evidence network for injectable on-label antibiotic regimes

The upper right-hand quadrant represents the estimated risk ratio, and the lower quadrant represents the 95% credible interval. Risk ratios are reported for registered antibiotic regimens only. Abbreviations are defined in Table 1.
Limitations of the body of work
The body of work that informed our literature review was large and relevant to the North American feedlot populations. In those characteristics, we believe that the studies met the important transit assumption of network meta-analysis: that the animals in a given study would be eligible for the treatments used in other studies. We did, however, find that lack of replication was an issue in some of the studies. For many interventions, only one trial was available. Therefore, estimates of effect sizes were often based on only one study. As with any scientific investigation, the value of replication is that it enables the consistency of effect measurements to be assessed.
Another limitation of the body of work is that the size of the effect of antibiotic use might change over time, and our time frame for study retrieval was not restricted. All methods of research synthesis assume that the network of evidence being used is static, meaning that the estimates presented in research reports are valid at the time the research was conducted and in subsequent years. However, there are a number of published studies suggesting that the frequency of antimicrobial resistance and the likelihood of multi-resistant organisms being present in host populations are growing (DeDonder and Apley, Reference DeDonder and Apley2015; Anholt et al., Reference Anholt, Klima, Allan, Matheson-Bird, Schatz, Ajitkumar, Otto, Peters, Schmid, Olson, McAllister and Ralston2017). The evaluation of trends over time in treated animals is complicated by the association between antibiotic treatment and the emergence of antimicrobial resistance in diagnostic samples (Magstadt et al., Reference Magstadt, Schuler, Coetzee, Krull, O'Connor, Cooper and Engelken2018). When antimicrobials, such as macrolides and tetracyclines, are used for both metaphylaxis and treatment, the emergence of resistant organisms becomes more likely (Timsit et al., Reference Timsit, Hallewell, Booker, Tison, Amat and Alexander2017a, Reference Timsit, Workentine, Crepieux, Miller, Regev-Shoshani, Schaefer and Alexander2017b). For those reasons, estimates of efficacy reported in older studies might no longer be valid because of recently emerging resistance, resulting in overestimations of current efficacy. Although it is unclear how the designation of ‘resistance’ empirically translates to metrics of clinical efficacy, most of the studies included in our meta-analysis were conducted early in the use period of the respective drugs and therefore represent the best-case scenario. If resistance is developing equally to all of the antibiotics, then the relative estimates of efficacy are likely still relevant. Alternatively, if resistance to some antibiotics is developing faster than that to other antibiotics, then the assumption of equal resistance development is not valid, and the relative estimates of efficacy have even more uncertainty than what is captured by research synthesis methods. One argument against changes in the prevalence of resistance being a major issue is that estimates derived from network meta-analyses very closely predict comparative efficacies in trials (O'Connor et al., Reference O'Connor, Coetzee, da Silva and Wang2013, Reference O'Connor, Yuan, Cullen, Coetzee, da Silva and Wang2016). Indeed, estimates of treatment efficacy for BRD from older studies were able to very accurately predict the comparative efficacies of new drugs (O'Connor et al., Reference O'Connor, Coetzee, da Silva and Wang2013, Reference O'Connor, Yuan, Cullen, Coetzee, da Silva and Wang2016). For example, for the comparison of gamithromycin (referent) and tulathromycin, the original model predicted a risk ratio of re-treatment of 0.54 (95% credible interval 0.27–0.87). In the subsequent randomized controlled trial, the observed risk ratio of re-treatment was 0.59 (95% confidence interval 0.45–0.78). For the gamithromycin (referent) and florfenicol comparison, the observed risk ratio in the randomized trial was 1.17 (95% confidence interval 0.83–1.64), and the estimated risk ratio from the prior model was 0.84 (95% credibility interval 0.48–1.3). For the gamithromycin and tilmicosin (referent) comparison, the risk ratio observed in the randomized trial was 0.99 (95% confidence interval 0.67–1.47), and the risk ratio estimated from the prior model was 1.09 (95% credible interval 0.64–1.79). If the data from older studies did not accurately represent current efficacy because of the emergence of resistant bacteria, those estimates would not be expected to be so close to the observed values. Assessment of error and bias is part of any good systematic review. The extent of random error is expressed by the credible intervals on the risk ratios and the rankings. The wide credible intervals in our results are mainly due to low replication within the studies. As shown in Fig. 2, for many antibiotic regimes, there is little publicly available information that could be incorporated into our meta-analysis. Large feedlot companies, veterinary consultants, and pharmaceutical companies likely have access to more trial data, but those data are not publicly available and could not be included in our meta-analysis.
The extent of possible systematic bias is presented individually in Table S9. It has been documented in a number of subfields of veterinary health that failure to randomize and blind studies is associated with biased estimates of intervention effects. However, in two previous systematic reviews and network meta-analyses related to BRD in beef cattle, we did not observe any overall changes in the magnitude of intervention effects (O'Connor et al., Reference O'Connor, Coetzee, da Silva and Wang2013, Reference O'Connor, Yuan, Cullen, Coetzee, da Silva and Wang2016). For some of the pairwise comparisons between antibiotics, Figs 4 and 5 show the relative contributions to the efficacy estimates of studies using different allocation approaches and blinding approaches (Papakonstantinou et al., Reference Papakonstantinou, Nikolakopoulou, Rucker, Chaimani, Schwarzer, Egger and Salanti2018). Our approach to those contribution matrices is that they should be used to interpret pairwise comparisons of particular interest. For example, the estimate for the comparison of OXY_1 and TMS (Table 4) suggests a 0.7-fold decreased risk of disease if OXY_1 is used instead of TMS. However, that estimate is based on several non-randomized studies, as indicated by the long red bar in Fig. 4, which also lacked information about blinding (Fig. 5). The problem with interpreting that kind of information is that, unlike in comparisons with placebos, we cannot hypothesize the direction of bias, because the contribution information is based on a pathway rather than a study. We therefore propose that the contribution information provides a visual indicator of the overall types of studies that contribute to a given estimate. Making more specific inferences about the direction of bias would require more knowledge about particular studies. We might also be skeptical about the results because of the sparse data, as indicated by the wide credible intervals. Although the data and derived estimates of efficacy represent the best evidence available, room for a healthy sense of uncertainty remains.
Limitations of the review process
Because our study was based on a systematic review, our first goal was to identify the relevant literature. The search was extensive, but it would have been ideal to have searched more databases if resources had permitted. Our database searches were complemented by other search approaches, such as hand searching. The database search strategies were sensitive, but they could have been made more sensitive by making greater use of the AND operator instead of the proximity operator, particularly to capture variant phrases related to the metaphylaxis concept. However, available resources dictated the use of the proximity operator since AND reduced precision and generated an unmanageably large volume of irrelevant records. It is also possible to search more websites and search engines, such as Google Scholar. However, within the limits of the available resources, we were able to achieve an extensive search.
Overall, the studies that contributed to the evidence base for our meta-analysis were well designed. Many studies reported randomization of cattle among treatment groups even if they did not provide evidence for the validity of the randomization. One of the major issues that we uncovered was differences in the baseline treatments that the cattle received when they arrived at the feedlot. Those differences could be interpreted in two ways: first, that the population was too heterogeneous to combine in a single analysis or, second, that the heterogeneity reflects the underlying population and is therefore a realistic representation of the cattle population most likely to receive antibiotics.
Conclusions
Overall, the existing body of research suggests that injectable antibiotics are generally effective at controlling BRD in feedlot cattle when they are administered at the time the cattle arrive at the feedlot. There is evidence that macrolides are the most effective injectable antibiotics for controlling BRD. The commonly used injectable oxytetracycline is also effective at controlling BRD, although less so than the macrolides. Oxytetracycline is already commonly used in groups of feedlot cattle to control, and treat BRD and therefore has advantages over the macrolides from an antibiotic stewardship perspective. Therefore, there might be an opportunity for wider use of injectable oxytetracycline to control BRD in feedlot cattle. If the use of antibiotics in feedlot cattle is discouraged, it is imperative that the efficacy of alternatives to antibiotics are fully understood so that producers can plan to mitigate the impacts that a lack of antibiotic use might have on animal welfare, health, and production.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S1466252320000031.
Acknowledgments
None to declare.
Author contributions
AOC developed the review protocol, coordinated the project team, assisted with the data analysis, interpreted the results, and prepared the manuscript drafts. DH conducted the data analysis, guided the interpretation of the results, commented on manuscript drafts, and approved the final version of the manuscript. ST conducted relevance screening, extracted data, provided guidance for the interpretation of the results, commented on the manuscript drafts, and approved the final manuscript. NS conducted relevance screening, extracted data, provided guidance for the interpretation of the results, commented on manuscript drafts, and approved the final manuscript. CWinder developed the review protocol, provided guidance for the interpretation of the results, commented on manuscript drafts, and approved the final manuscript version. JG developed the review protocol, provided guidance on the creation of the search, commented on manuscript drafts, and approved the final manuscript. HW developed the review protocol, developed and conducted the search, commented on the manuscript drafts, and approved the final version of the manuscript. BWang developed the review protocol, provided guidance on the conduct of the analyses and interpretation of the results, commented on the manuscript drafts, and approved the final manuscript version. CWang developed the review protocol, provided guidance on the conduct of the analyses and interpretation of the results, commented on manuscript drafts, and approved the final manuscript version. BWhite developed the review protocol, provided guidance on the conduct of the analyses and interpretation of the results, commented on manuscript drafts, and approved the final manuscript version. RL developed the review protocol, provided guidance on the conduct of the analyses and interpretation of the results, commented on manuscript drafts, and approved the final manuscript version. JS developed the review protocol, provided guidance for the interpretation of the results, commented on manuscript drafts, and approved the final manuscript version.
Financial support
Support for this project was provided by The Pew Charitable Trusts.
Conflict of interest
JS, CW, NS, CW, DH, JG, HW, and BWang have no conflicts to declare. AOC and ST have conducted prior reviews for Bayer Animal Health which may manufacture one or more of the products assessed. RL has conducted research or consulting for Zoetis Animal Health, Merck & Company, CEVA Biomune, Boehringer Ingelheim Vetmedica, and Merial Animal Health which may manufacture one or more of the products assessed. BW has conducted research or consulting for Bayer Animal Health, Boehringer Ingelheim, Elanco Animal Health, Merck Animal Health, Merial Animal Health, and Zoetis Animal Health which may manufacture one or more of the products assessed.
Publication declaration
The authors declare that this is a full and accurate description of the project and that no important information or analyses are omitted.











