Effective conservation management requires identification of the population's genetic structure, and genetic distinctness of a population has long been recognized as a key issue for conservation concern (Moritz, Reference Moritz1994). It is important, however, that arguments for genetic distinctness are based on scientifically valid information so that the efficiency of management efforts is not compromised. Here we consider the situation where genetic distinctness is used as a conservation argument but the scientific evidence appears weak: the harbour porpoise Phocoena phocoena of the Baltic Sea. The harbour porpoise is considered threatened in parts of its distribution range and is categorized as Vulnerable on the IUCN Red List (IUCN, 2007). It is the only cetacean occurring regularly in the Baltic Sea and in the adjacent Danish Belt, Skagerrak and Kattegat Seas (Berggren & Arrhenius, Reference Berggren and Arrhenius1995; Fig. 1).
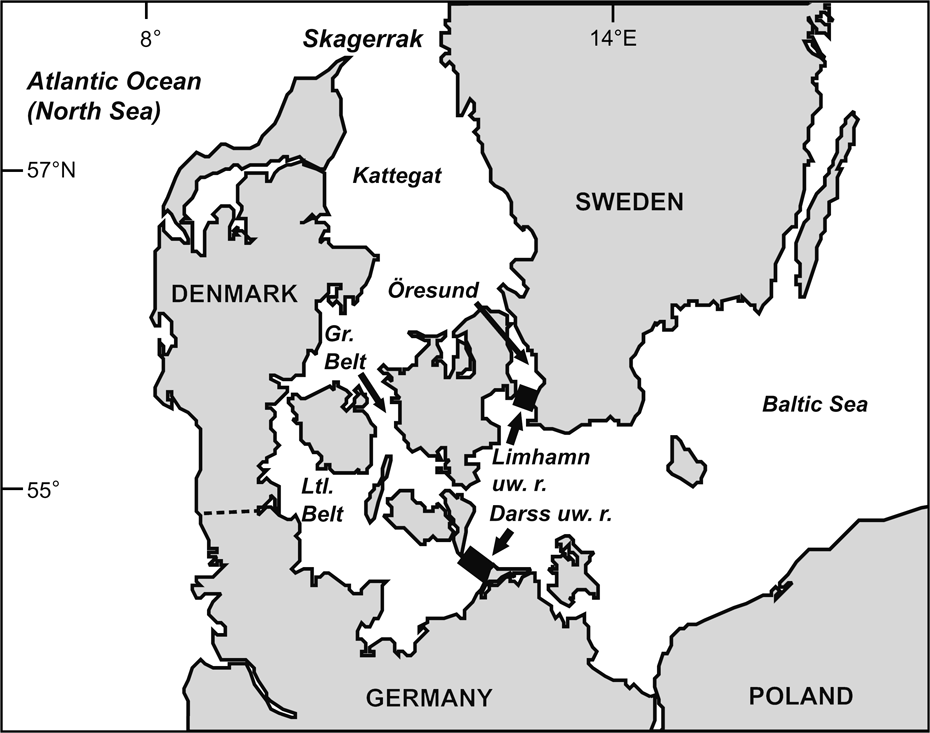
Fig. 1 The Baltic Sea and other water bodies discussed. The western borders of the Baltic proper are defined by the Limhamn and Darss underwater ridges (uw. r.).
Conservation concern (ABDG, 2001; ASCOBANS, 2002; Lindahl et al., Reference Lindahl, Westerberg and Rappe2003) for the harbour porpoise of the Baltic Sea proper (defined by the Limhamn and Darss underwater ridges; Fonselius, Reference Fonselius1995; IWC, 2000; Fig. 1) is based on the conjectures that the population is declining, comprises <500 animals (Berggren & Arrhenius, Reference Berggren and Arrhenius1995; Hiby & Lovell, Reference Hiby and Lovell1996; Berggren, Reference Berggren2003), and is genetically distinct and reproductively isolated from adjacent populations west of the Baltic, i.e. in the Belt, Kattegat and Skagerrak Seas (IWC, 2000). The population estimate for these latter regions is c. 40,000 animals (Hammond et al., Reference Hammond, Berggren, Benke, Borchers, Collet and Heide-Jørgensen2002).
The notion of a genetically distinct Baltic population is widely accepted by many involved in the management of the species (ASCOBANS, 2002; Huggenberger et al., Reference Huggenberger, Benke and Kinze2002; Koschinski, Reference Koschinski2002; Teilmann et al., Reference Teilmann, Dietz, Larsen, Desportes, Geertsen and Andersen2004; HELCOM, 2006). For instance, a political agreement on the Conservation of Small Cetaceans of the Baltic and North Seas (ASCOBANS), ratified under the Bonn Convention by nine north European countries, largely focuses on the Baltic harbour porpoise. Considerable resources are allocated annually by the Swedish authorities to promote the conservation of harbour porpoise in Swedish waters. The Baltic porpoise is classified as ‘a vulnerable geographical population' (Ingelög et al., Reference Ingelög, Andersson and Tjernberg1993; ASCOBANS, 2002), and suggestions for increased research efforts imply a genetically distinct Baltic population (Hopkins, Reference Hopkins2005).
The scientific support for a genetically distinct Baltic harbour porpoise primarily refers to Wang & Berggren (Reference Wang and Berggren1997) and Tiedemann et al. (Reference Tiedemann, Harder, Gmeiner and Haase1996). We suggest that neither provides strong evidence for such a claim. Both studies are based on mitochondrial DNA (mtDNA). Reflecting only the female genetic structuring, neither study provides insight into the variation in nuclear genes, which contain the vast majority of the genetic information. Tiedemann et al. (Reference Tiedemann, Harder, Gmeiner and Haase1996) compared haplotypes of 20 porpoises collected in the southern Baltic and Belt Seas area (Fig. 1) with those of 19 specimens from the North Sea, obtaining a statistically significant frequency difference (P <0.01). However, the sample referred to as Baltic also appears to have included porpoises from the Belt Seas, and this study does not therefore strictly test for genetic distinctness of a Baltic porpoise.
Wang & Berggren (Reference Wang and Berggren1997) performed a strict comparison of samples from the Baltic proper versus animals collected in the Kattegat-Skagerrak area (Table 1) and reported a statistically significant difference of haplotype frequency distributions (using option Monte of the Restriction Enzyme Analysis Package, REAP v. 4.1; McElroy et al., Reference McElroy, Moran, Bermingham and Kornfield1992). However, our recalculations do not support the notion of statistical significance. Using the data of Wang & Berggren (Reference Wang and Berggren1997) we reanalysed the haplotype frequency difference between the Swedish Baltic and the Kattegat-Skagerrak areas, where they report a P of 0.035. We used several statistical packages (including option Monte in REAP v. 4.1), applying both exact calculations and simulation approaches, but did not obtain P <0.05 (P-values were 0.08 – 0.22; Table 2).
Table 1 Observed haplotype frequencies of Phocoena phocoena from the Baltic Sea proper (see text for definition) and from the Kattegat-Skagerrak Seas (Fig. 1; data from Wang & Berggren, Reference Wang and Berggren1997).
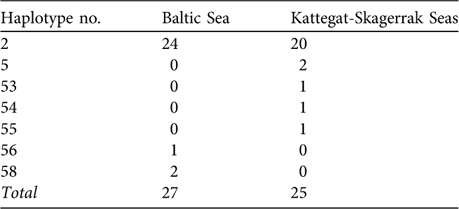
Table 2 Results of reanalyses for equal haplotype frequencies of Phocoena phocoena (data of Table 1) from the Baltic vs the Kattegat-Skagerrak Seas (Fig. 1) using four software packages.
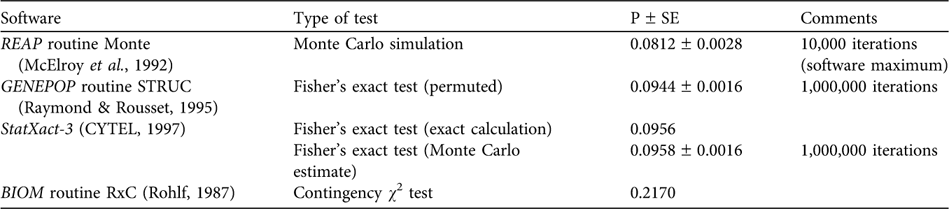
Clearly, the difference between a P of 0.035 and most of our recalculated values is not large, and strict application of the P = 0.05 limit for defining statistical heterogeneity should be exercised with caution. Nevertheless, when applying this standard criterion for statistical significance the data of Wang & Berggren (Reference Wang and Berggren1997) do not support the idea of a Baltic population that is genetically distinct from that of the Kattegat-Skagerrak area.
There are at least four additional molecular genetic studies of harbour porpoises in the Baltic-Belt-Kattegat-Skagerrak region but none of them address the issue of a genetically distinct Baltic population (Andersen, Reference Andersen1993; Andersen et al., Reference Andersen, Holm, Clausen, Kinze, Blix, Walløe and Ulltang1995, Reference Andersen, Holm, Siegismund, Clausen, Kinze and Loeschcke1997, Reference Andersen, Ruzzante, Walton, Berggren, Bjørge and Lockyer2001), and the definition of the geographic range of the Baltic Sea varies considerably (Palmé et al., Reference Palmé, Laikre and Ryman2004). The consensus from these four studies is that some general structuring exists within the region examined but the pattern has not been identified, and distinct population segments have not been recognized (Palmé et al., Reference Palmé, Laikre and Ryman2004). Over all studies, only 38 specimens from the Baltic Sea have been genotyped.
The degree of genetic divergence among harbour porpoises within the Baltic-Belt-Kattegat-Skagerrak appears to be low. Using the data and sample sizes (n = 27 and 25) of Wang & Berggren (Reference Wang and Berggren1997; Table 1) we obtain an estimate of FST (the proportion of the total genetic diversity attributable to differences between populations) between the Baltic and the Kattegat-Skagerrak of 0.007 (implying that only 0.7% of the total gene divergence is due to differences between regions). We used the software POWSIM (Ryman & Palm, Reference Ryman and Palm2006) to simulate 1,000 FST values from two samples drawn from the same population (FST = 0) and found that, with these sample sizes, the probability of obtaining an FST ≥0.007 is c. 0.30. Therefore, this observation is not unlikely even with complete random mating (panmixia) among these waters.
Palsbøll et al. (Reference Palsbøll, Bérubé and Allendorf2007) suggested that rejection of panmixia should not constitute the primary basis for identification of conservation units. Rather, the degree of demographic independence and genetic divergence constitute the criteria for assigning such units. They suggest that with 10% or more exchange of migrants, two populations should be considered demographically connected and therefore managed jointly. With lower levels of exchange populations may be regarded as separate management units. The degree of migration between porpoises in the Baltic and those further west is unknown. However, assuming an effective population size of 200 in the Baltic (Hiby & Lovell, Reference Hiby and Lovell1996; Berggren, Reference Berggren2003), that female and male effective population sizes and migration rates are equal, and migration-drift equilibrium (cf. Birky et al., Reference Birky, Maruyama and Fuerst1983), FST for mtDNA will be 0.05 with 20 migrants per generation (i.e. 10% migration). Using the same simulation approach as earlier we estimate the probability of obtaining the observed FST = 0.007 or lower as 0.35. Therefore, with these haplotype frequencies and sample sizes it is not possible to distinguish a situation of panmixia from that with a 10% exchange.
No other FST estimate between Baltic porpoises and those further west is available. However, Andersen et al. (Reference Andersen, Ruzzante, Walton, Berggren, Bjørge and Lockyer2001) report an FST of 0.005 between inner Danish waters (including the Baltic, Belt and Kattegat Seas) and the Skagerrak/Danish North Sea. This low estimate indicates considerable genetic exchange between populations in this region.
Confidence in conservation genetic arguments is undermined when supporting evidence is weak. Strong opposition to protective measures almost always arises where, as in the case of the Baltic harbour porpoise, fisheries are requested to change methods and gear to protect a species. Using genetic justification for protection that may later have to be reconsidered could undermine the confidence in conservation genetics. There may be other grounds for conserving particular groupings (Taylor & Dizon, Reference Taylor and Dizon1999), and such may exist for the Baltic porpoise, but here we are primarily concerned with the use of genetic arguments without scientific support.
A thorough investigation of the pattern of genetic substructuring of the harbour porpoises of this region is required. There are three basic types of spatial genetic differentiation: distinct local populations, continuous genetic change, or lack of differentiation (Laikre et al., Reference Laikre, Palm and Ryman2005). The management and protection required differs between these types and would also be affected by the geographic location of possible genetic groupings. If the Baltic harbour porpoise represents the same population as that of the Belt and Kattegat Seas there is no obvious genetic argument for the strong conservation efforts currently employed in the Baltic. But, if the Baltic porpoises represent a reproductively isolated population or considerable part of a cline, then strong conservation action is needed.
Acknowledgements
We thank Per J. Palsbøll for valuable discussions and suggestions, and Liselotte W. Andersen and two anonymous reviewers for valuable suggestions. Financial support from the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (to LL), The Swedish Research Council (to NR and LL), and the Swedish Environmental Protection Agency (to NR) is acknowledged.
Biographical sketches
Anna Palmé's research focuses on spatial genetic structure over micro-geographical areas in the absence of barriers to migration. Linda Laikre, Fred Utter and Nils Ryman share an interest in conservation genetics and the genetic effects of the human exploitation of natural populations. Linda Laikre also focuses on issues related to monitoring such effects, and Fred Utter's and Nils Ryman's research deals with the genetic structuring of natural populations and the processes that create these patterns.





