INTRODUCTION
Influenza viruses are in the family Orthomyxoviridae and are distributed in three genera, influenza A, B and C. Influenza A and B cause substantial morbidity and mortality in humans with influenza A viruses causing greater mortality in most epidemic years. Occasional antigenic shift within influenza A viruses results in significant human disease, leading to influenza pandemics. It has been reported that between 3 and 5 million cases of severe influenza disease occur each year [Reference Nicholson, Wood and Zambon1]. Annually, influenza-associated mortality has been estimated to range between 500 000 and 1 000 000 cases [Reference Russell2–Reference Flahault4].
Influenza A and B viruses contain two major surface glycoproteins, haemagglutinin (HA) and neuraminidase (NA). Influenza A viruses are classified into distinct subtypes based on their HA and NA phenotypes. Sixteen HA subtypes in combination with nine NA subtypes have been identified in aquatic birds [Reference Fouchier5, Reference Hampson and Mackenzie6]. Currently, the predominant influenza virus subtypes circulating in human populations are influenza A(H1N1) and influenza A(H3N2) [Reference Nelson7, Reference Sampath8]. Selective pressures imparted on NA and HA glycoproteins results in ‘antigenic drift’ thereby allowing influenza viruses to evade the host's immune responses [Reference Katz9, Reference Williams10].
Identification and subtyping of influenza viruses are routinely performed using PCR [Reference Zhang and Evans11, Reference Wright12]. The relationship between influenza viruses can be determined through antigenic and genetic comparative studies [Reference Davydov13]. Specific influenza virus clades have been described for both influenza A and B viruses based on nucleotide sequencing and phylogenetic characterizations of the HA gene [Reference Pechirra14–Reference Martin18].
Global strategic plans to address and reduce seasonal and pandemic influenza require, in part, unbiased epidemiological estimates in addition to the characterization of regional influenza viruses recovered during endemic and epidemic surveillance [Reference Flahault4]. Surveillance programmes are useful in monitoring the temporal and geographical changes in circulating influenza viruses in any given region. Influenza surveillance is especially critical in SE Asia as it has been reported that this region appears to be the source which ‘seeds’ the subsequent annual global seasonal epidemics [Reference Russell2].
Our laboratory established hospital-based surveillance to ascertain the aetiologies contributing to acute undifferentiated fever in rural Cambodian patients. This report describes the epidemiology of influenza from acutely ill Cambodian patients seeking healthcare from December 2006 to December 2008.
MATERIALS AND METHODS
Site selection and study enrolment
Hospital-based fever surveillance was initiated in December 2006. Patients were initially recruited from two referral hospitals, but during the course of the study, additional healthcare facilities were added: two in August 2007, one in October 2007, one in December 200, one in February 2007, one in March 2008 and one in April 2008, giving an overall total of nine. Five of the healthcare sites were located in operational district A (peri-urban); four healthcare sites were located in operational district B (rural). Both healthcare sites were located within 50 km of Phnom Penh in south-central Cambodia.
Patients were clinically evaluated by a physician or medical assistant and recruited for study participation if they met inclusion criteria: at least 24 h of fever (tympanic membrane temperature >38·0°C), were aged ⩾2 years, and after medical examination, had no obvious source of external wound infection. Subsequently, the physician or medical assistant in each clinic obtained written informed consent, administered a pretested enrolment questionnaire, and performed a medical examination. Laboratory technicians collected clinical specimens (blood, sputum and stools, as indicated). All study subjects were requested to return 14–21 days after the initial clinic visit for a follow-up medical visit. During the follow-up, a questionnaire was completed and a blood specimen collected.
Definitions
Influenza-like illness (ILI) was defined by fever (>38·0°C), plus two of the following symptoms: cough, sore throat, headache, myalgia, and rhinorrhoea [Reference Boivin19–Reference Nicholson21]. Diarrhoea was defined as ⩾3 loose or liquid stools in any 24-h period or ⩾2 loose or liquid stools and an associated symptom common to diarrhoeal disease (i.e. abdominal cramping, abdominal pain, fever, nausea, vomiting). Recent travel was defined as the individual leaving their respective home for at least five consecutive days within the 2 months preceding enrolment. Seasonality was defined into two categories, rainy (May–October) and dry (November–April) [Reference Vong22]. Operational district is a designation used by the Cambodian government to denote a continuous geographic region under the purview of a single government office.
Specimen collection
For each enrolled patient, we collected blood and one throat and one nasal swab. Standard venepuncture procedures were used to collect all blood specimens. For nasal swabs, a dry polyester swab was inserted into the nostril, parallel to the palate, slowly withdrawn and placed in a vial containing 2–3 ml virus transport medium (VTM). For throat swabs, both tonsils and the posterior pharynx were swabbed vigorously, and the swab placed in 2–3 ml VTM. All inoculated vials were kept at 4°C until transported to our laboratory. All specimens were received between 24 h and 72 h after collection. Clinical specimens found to be positive for influenza by real-time reverse-transcriptase polymerase chain reaction (rRT–PCR) were shipped to the WHO Collaborating Center for Influenza at the Centers for Disease Control and Prevention (CDC) in Atlanta, Georgia, USA for virus isolation, genetic characterization and antiviral testing.
rRT–PCR amplification
Ribonucleic acid (RNA) was extracted from nasal and throat swabs using QIAamp viral RNA mini kits (Qiagen, Germany) according to the manufacturer's instructions and stored at −70°C. Influenza virus genome was detected using a reverse real-time PCR assay developed to detect influenza A and B viruses as well as influenza A viruses of H1, H3 and H5 subtypes. Real-time assays were developed at the CDC (assays and primer sequences are available upon request under a material transfer agreement). One-step Rrt–PCR was performed in a final volume of 25 μl containing 5 μl extracted RNA, 12·5 μl buffer mix and 0·5 μl Superscript III/Platinum Taq-Enzyme mix, 20 U RNAse-out (Invitrogen, USA), 0·8 μm for each primer and 0·2 μm of probe. The Rotor-Gene 6000 real-time thermocyler (Corbett Life Science, Australia) was used for all PCR reactions. The thermocycling parameters for all targets consisted of 50°C for 30 min, 95°C for 2 min, and 45 cycles with 95°C for 15 s, 55°C for 30 s (CDC).
Virus isolation
Virus isolation was conducted at the Influenza Division, CDC (USA) using 9- to 11-day-old embryonated eggs or Madin–Darby canine kidney (MDCK) cells as described previously [Reference Kendal and Cate23].
Haemagglutination inhibition (HI) assay
Virus isolates were characterized by HI tests using post-infection ferret sera with turkey erythrocytes as previously described [Reference Kendal and Cate23].
Influenza nucleotide sequence analysis
Influenza viruses isolated from clinical specimens were used for sequencing of the HA gene. Viral RNA extraction was performed using the Qiagen BioRobot M48 workstation with the MagAttract Viral RNA M48 kit (Qiagen, USA). RNA was used as template for RT–PCR with subtype and segment-specific oligonucleotide primers. Reaction conditions and primer sequences are available from the authors upon request. Nucleotide sequencing reactions were performed with Big Dye Terminator chemistry version 3.1 (Applied Biosystems, USA) and resolved on an ABI 3730xl genetic analyser (Applied Biosystems). Sequence data was assembled and sequences were generated using Sequencher™ version 4.7 software package (Gene Codes Corporation, USA). All sequence data used in this study are available from GenBank (accession numbers FJ375205–FJ375208).
Phylogenetic analysis
Phylogenetic trees were created using the neighbour-joining method, nucleotide model of Kimura two-parameters, in MEGA version 4 [Reference Tamura24]. In addition, a subset of H1 HA gene sequences available in the public domain was included for comparative purposes in the analyses.
Antiviral drug susceptibility assays
Susceptibility of the viruses to oseltamivir and zanamivir was assessed using the chemiluminescent NA inhibition assay which utilizes a 1,2-dioxetane derivative of sialic acid as substrate. The assay was performed using the NA-Star® kit (Applied Biosystems) as described by Sheu et al. [Reference Sheu25]. Calculation of 50% inhibitory concentration (IC50) values and curve-fitting were performed by Robosage version 7.31 software (GlaxoSmithKline, USA), an add-in for MS Excel (Microsoft Corp., USA) using the equation y=V max×[1 – (x/(K+x))] as previously described [Reference Sheu25, Reference McKimm-Breschkin26].
Pyrosequencing to detect molecular markers of resistance to adamantanes was performed as previously described [Reference Bright27, Reference Deyde28] using the PSQ96 Sample Preparation kit and the PSQ96 SQA Reagent kit (Biotage AB, Sweden) on the PSQ96MA platform. Pyrosequence data consisting of 45–60 nucleotide reads were quantified and background-corrected using PSQ96MA version 2.0.2 software (Biotage AB). Sequences were aligned and analysed using DNAStar analysis programs (DNAStar, USA).
Statistical analysis
All data were double-data entered into MS Access (Microsoft Inc.). Categorical data were analysed by the use of either χ2 test (expected cell frequency >5) or Fisher's exact test (expected cell frequency ⩽5). Continuous data were assessed for normality and if normally distributed, parametric statistics was used. If the data were non-normally distributed, then non-parametric testing was used. Data were imported into SAS version 9.1 (SAS, USA), which was used for all statistical analyses. All statistical tests were two-tailed and significance was defined as P<0·05.
Ethical considerations
Eligible subjects voluntarily enrolled in accordance with an Institutional Review Board protocol approved by U.S. NAMRU2 and the National Ethics Committee of the Royal Kingdom of Cambodia, Ministry of Health.
RESULTS
Demographic and clinical findings
From December 2006 to December 2008, 4451 patients were enrolled (Table 1). Of patients who had a clinical specimen tested for influenza (n=4233), the median age was 11 years [interquartile range (IQR) 6–25 years], 54·7% (n=2314) were males, with roughly equal numbers enrolled from each field site. Most of the participants were treated as outpatients (n=3864, 91·5%). The median duration of fever at enrolment was 3 days (IQR 2–4 days). The five most common symptoms reported by patients were headache (n=2713, 64·6%), cough (n=2340, 55·5%), sore throat (n=1908, 45·5%), chills (n=1815, 43·1%), and malaise (n=1732, 41·4%). When patients were asked if they knew someone with similar symptoms, 404 (9·8%) indicated they did with >98% reporting a sick household member or neighbour.
Table 1. Demographics of Cambodia patients presenting with febrile illness stratified by influenza status

Missing data from each covariate accounts for percentage differences.
Of the 4233 patients, 1151 (27·2%) were PCR positive for influenza (see Tables 1 and 2). Influenza infections were high during the consecutive months of August (n=136, 11·8%), September (n=330, 28·7%), and October (n=420, 36·5%), while the months of February (n=3, 0·26%), March (n=0), and April (n=1, 0·09%) were the lowest (see Table 1 and Fig. 1). The proportion of influenza-positive patients was similar for males (n=636, 55·3%) and females (n=515, 44·7%, P=0·6), but decreased with increasing age (β=−0·04, P<0·0001). The median age of influenza-positive patients was 8 years (IQR 5–13) while the median age of influenza-negative patients was 13 years (IQR 6–28) (Kruskal–Wallis, P<0·0001) (Table 1). There was an initial increase in influenza positivity from just over 30% in children aged 2–5 years to almost 40% in those aged 6–10 years, which then decreased across the remaining age categories (Fig. 2). Influenza was detected at a higher rate in outpatients (n=1112, 28·8%) compared to hospitalized patients (n=35, 9·8%). Persons with influenza were slightly more likely to report knowing someone with similar symptoms compared to febrile patients who were influenza negative, 12·4% compared to 8·8% (P=0·006), respectively. We noted slight, albeit statistically significant, differences in the identification of patients across field sites, with operational district A at 48·7% and operational district B at 51·3% (P<0·0001).

Fig. 1. Monthly distribution of enrolment (□) and influenza positivity (▪) in patients seeking healthcare for febrile illness in south-central Cambodia from December 2006 to December 2008.

Fig. 2. Influenza positivity by age category stratified by gender in patients seeking healthcare for febrile illness in south-central Cambodia from December 2006 to December 2008.
Table 2. Influenza cases by virus subtypes
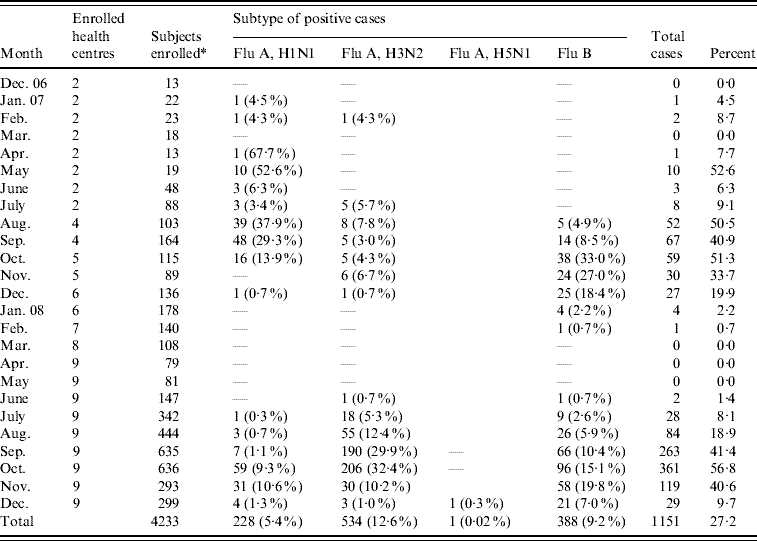
* Swabs not taken for nine enrollees.
Symptoms statistically associated with influenza-positive vs. influenza-negative cases included cough (68·8% vs. 50·5%, P<0·0001) and sore throat (55·0 vs. 41·9%, P=0·0004) (Table 1). Malaise (33·3% vs. 44·4%, P<0·0001), chills (38·3% vs. 44·8%, P<0·0001), muscle aches (18·1% vs. 24·6%), joint pain (11·5% vs. 18·3%), diarrhoea (3·9% vs. 7·2%, P<0·0001) and abdominal cramping (17·7% vs. 22·4%, P<0·0001) were more common in influenza-negative patients. No statistically significant difference was noted for headache, nausea or vomiting in influenza-positive patients compared to influenza-negative patients.
Of 1151 influenza viruses detected, 228 (19·8%) were influenza A(H1N1), 534 (46·4%) were influenza A(H3N2) and 388 (33·7%) were influenza B (Table 2). In addition, we detected a single influenza A(H5N1) strain during December 2008 (Table 2). The data suggested three distinct temporal patterns: influenza A(H1N1) occurred in successive months from April to October 2007, influenza A(H3N2) from July to December 2007, and influenza B from August to February 2008. No influenza cases were detected from March 2008 to May 2008, but cases reappeared starting in June 2008 for all subtypes (Fig. 3).

Fig. 3. Influenza subtype distribution by date. Specimens from Cambodian patients reporting for primary medical care with subjective fever.
On 2 December 2008, a 19-year-old male from Kandal Province was enrolled in the study with complaints of fever, malaise, myalgia, headache, sore throat and cough over the previous 8 days. Subsequent laboratory testing by rRT–PCR, identified avian influenza A(H5N1) virus. A diagnosis of influenza A(H5N1) was confirmed by the Pasteur Institute of Cambodia on 12 December 2008 and the case was officially recognized as the eighth avian influenza human infection recorded in Cambodia by the WHO (P. Buchy, personal communication, 2008). The patient was hospitalized at Callette Hospital in Phnom Penh, treated with oseltamivir and subsequently released after 3 weeks hospitalization. The victim reported eating an infected chicken prior to the onset of symptoms (P. Buchy, personal communication, 2008).
Antigenic analysis
Viruses collected in 2007 were characterized at the WHO Collaborating Center for influenza at CDC (USA). HI assays using post-infection ferret antisera revealed that the majority of influenza A(H1N1) viruses identified in our study were antigenically related to A/Solomon Islands/03/2006, the influenza A(H1N1) vaccine component for the 2007–2008 influenza season, or to the antigenically related A/Hong Kong/2652/2006 reference strain. All the influenza B viruses tested belonged to the influenza B/Victoria lineage and were closely related to the B/Malaysia/2506/2004 reference strain (data not shown). Influenza A(H3N2) viruses tested were antigenically distinguishable from the 2007–2008 vaccine strain A/Wisconsin/67/2005, but were closely related to the 2008–2009 vaccine strain A/Brisbane/10/2007 (data not shown).
Genetic analysis of HA1 of influenza A(H1N1)
In the phylogenetic tree of the HA gene of influenza A(H1N1) viruses, two major clades were evident with clade 2 separated into distinct subgroups A–C (Fig. 4). Viruses isolated in Cambodia clustered together within the 2C subgroup. The 2C subgroup viruses shared amino-acid changes in HA1 at positions 45 (serine to asparagine), 90 (lysine to arginine), 192 (arginine to methionine), 193 (alanine to threonine) and 197 (threonine to lysine). Subgroup 2C viruses from Cambodia were found to be co-circulating during the same time period with similar viruses detected in the USA, Australia, and SE/E Asia.

Fig. 4. Evolution of the HA of A(H1N1) viruses from Cambodia sampled during 2007–2008 season. Phylogenetic tree was constructed from the HA1 genes of viruses sequenced during the study (n=4) and 36 viruses with HA sequences available in GenBank. The two major genetic clades and subclades designated 1 and 2A–C were identified. Cambodian isolates are in boldface. Characteristic amino-acid changes are shown at the appropriate nodes.
Antiviral resistance
Using the chemiluminescent NA inhibition assay, all the tested influenza A(H1N1) (n=20), influenza A(H3N2) (n=7) and influenza B (n=2) viruses were sensitive to oseltamivir and zanamivir, while M2 gene pyrosequencing data revealed that all A(H1N1) (n=16) and A(H3N2) (n=7) viruses were resistant to adamantanes and had the S31N mutation in their M2 protein.
DISCUSSION
The results presented here highlight three main findings: (1) influenza A and influenza B infections contribute to significant morbidity in patients seeking treatment at healthcare facilities in south-central Cambodia, (2) data obtained from 1511 laboratory-confirmed influenza cases clearly demonstrate a seasonality, (3) passive hospital-based surveillance of acute febrile illnesses can serve as a sentinel for emerging infectious threats such as the avian H5N1 influenza virus.
Upon stratification by influenza results, we found no statistical difference when comparing males and females. We found that only 2% of the influenza-positive patients were hospitalized during enrolment, while for influenza-negative patients, admission rates reached almost 10%. Supporting laboratory studies identified other infectious aetiologies (e.g. dengue, malaria, etc.) probably contributing to the increase in admissions of our influenza-negative patients. Of interest was the finding that just over 12% of our influenza-positive patients reported knowing someone with the same symptoms, of which >90% were noted in family members and/or neighbours. However, in the influenza-negative group, nearly 9% reported knowing someone with the same symptoms. Albeit statistically significant, these estimates may be misleading as the villages in which these patients reside are small it is likely that everyone will be aware of the general health status of all community members. Although influenza viruses are thought to be highly infectious and frequently spread among family members and neighbours, our finding is consistent with our understanding of the epidemiology of influenza [Reference Cox and Subbarao29].
In addition to fever, many patients presented with other symptoms commonly associated with respiratory disease such as cough (55·5%), headache (64·6%) and sore throat (45·5%). Our results showed that influenza infection was statistically associated with cough and sore throat, but not with other symptoms. This analysis supports the current ILI case definition established by WHO guidelines.
Overall, nearly 50% of our study group were children aged ⩽10 years. This finding is not surprising since it has been reported that children tend to harbour many pathogens, including respiratory viruses [Reference Navarro-Mari20]. In Cambodia, we observed that influenza contributed to nearly 25% of all-age acute febrile illnesses during 24 months; in children the influenza prevalence rate increased to 35%. Further analyses found an inverse relationship between age and the identification of influenza. The highest influenza rates were noted for the youngest age groups, children aged <10 years. The rates decreased into the adult groups and remained steady around 10%. It is probable that acquired influenza immunity could explain the decrease; however, there are other possible explanations: The incidence of other fever-inducing agents increases with age and thereby changes the pattern of acute infection. As a result, adults may seek treatment for influenza elsewhere or have less severe symptoms not requiring medical attention with an increase in self-medication for fever and influenza-like symptoms. Moreover, possible socioeconomic behaviours might alter healthcare-seeking in adults.
There appeared to be a seasonality associated with influenza in our study group, with the months of August, September and October as the ‘high’ season, coinciding with the rainy season. We noted a parallel increase in the number of patients seeking healthcare for fever and subsequent confirmation of influenza infection starting in June 2007, peaking September/October 2007 and then rapidly decreasing up to January 2008. This increase was noted to coincide with the end of rainy season in Cambodia, which typically starts in June and lasts until October. Data in our study were acquired over 24 months and we cannot exclude the possibility that longer surveillance would identify additional seasonal patterns. However, the apparent increase in influenza infections during Cambodia's rainy season is similar to the seasonal pattern observed during the winter months in temperate regions such as Europe and North American [Reference Nelson7, Reference Finkelman30–Reference Viboud32]. The observed influenza seasonality in our study parallels the regional influenza seasonality in neighbouring countries as well as other countries on or near the same latitude [Reference Nelson7, Reference Viboud, Alonso and Simonsen31, Reference Park and Glass33–Reference Hsieh36], although these regions have a reduced seasonal influenza epidemic magnitude when compared to temperate regions. In neighbouring Thailand, up to 35% of ILI outpatients may be positive for influenza during the months of May–September; whereas smaller peaks are often observed in the cooler, dry months, December–February (M. Simmerman, personal communication, 2008). Many other countries in SE/E Asia report sporadic seasonal epidemics throughout any given year, but in general there is a consistent increase in influenza prevalence during the winter months or rainy seasons and a low level of influenza disease in the summer or dry months. In a survey of influenza viruses collected between 2003 and 2006 in Taiwan [Reference Jian37], A(H3N2) viruses demonstrated year round influenza activity, while influenza B and A(H1N1) cases peaked in the winter months [Reference Jian37, Reference Lin38]. Our work in Indonesia (2004–2008) has shown that continuous year-round transmission of both influenza A and B viruses occurs (P. Blair, personal communication) with temporal dynamics similar to other regional countries' influenza surveillance reports. The Indonesian data demonstrate rainy season peaks for both influenza A and B, with a significant decrease during the dry months. In Singapore, influenza viruses circulate year round, with a slight increase in the magnitude of a bimodal epidemic of influenza infections observed in April–July and November–January [Reference Shek and Lee39–Reference Chew41].
SE/E Asia has been the epicentre for the previous emergence of novel respiratory viral infections as evidenced by influenza A(H5N1) in 1997 [Reference Claas42] re-emerging in 2003 [Reference Peiris43] in Hong Kong, and the SARS-CoV Hong Kong outbreak in 2003 [Reference Liang44]. Since 1997, 407 human influenza A(H5N1) cases have been reported from 15 nations with 254 deaths [case-fatality rate (CFR) of 62%] (http://www.who.int/csr/disease/avian_influenza/en/), including seven cases in Cambodia (100% CFR). The first six Cambodian victims were infected with clade 1, genotype Z H5N1 viruses [Reference Buchy45]. A report by Vong et al. [Reference Vong22] found evidence of influenza A(H5N1) RNA from several environmental samples collected from areas where recent influenza A(H5N1) cases (bird or human) had occurred in Cambodia [Reference Vong22]. The 19-year-old patient identified in our febrile illness study and subsequently diagnosed with influenza A(H5N1), was the eighth human infection in Cambodia and the first case that did not result in fatality (Cambodia Ministry of Health, 2008, personal communication). As has been reported for most avian influenza A(H5N1) cases, the probable source of the virus was direct contact with infected poultry [Reference Vong22, Reference Brankston46]. Interestingly, upon medical examination, the patient met the study enrolment criteria, but was not considered a probable avian influenza case until our laboratory identified influenza A(H5N1) by rRT–PCR. Specimens were confirmed by Pasteur Institute, Cambodia (P. Buchy, personal communication, 2008). While the misdiagnosis probably reflects the low frequency of avian influenza cases seen by practising physicians in rural Cambodia, it underscores the necessity of providing ongoing medical and public health educational programmes aimed at improving the diagnostic recognition of possible avian influenza disease.
Influenza infections are the cause of significant morbidity and mortality worldwide, with seasonal influenza afflicting the very young and the old. Reports estimate that worldwide between 3 and 5 million cases of severe illness and nearly 500 000 deaths each year are attributed to influenza [Reference Nicholson, Wood and Zambon1]. ILI surveillance programmes are useful for monitoring the temporal and geographical changes in circulating influenza viruses from year to year and across distinct geographic regions. Influenza surveillance is especially critical in SE/E Asia as it has been reported that this region appears to be the source which ‘seeds’ the subsequent annual global seasonal epidemics [Reference Russell2]. The identification and recovery of avian influenza A(H5N1) has pushed the global medical community to implement several new influenza surveillance programmes. Our results indicate that in south-central Cambodia, influenza virus circulates throughout most the year with an apparent seasonal peak toward the end of rainy season.
In this work, we describe the epidemiology of influenza in patients seeking healthcare for febrile illness in Cambodia from December 2006 to December 2008. Published reports support the hypotheses that continual intra-regional circulation of influenza virus in SE/E Asia ‘seeds’ annual global influenza epidemics seen in North American, Europe, Africa and South America. Based on our results, as well as other regional reports, integrated influenza surveillance in SE/E Asia is critical for regional estimates of the burden of influenza/respiratory disease, identification of circulating influenza strains, monitoring of antiviral resistance, providing rapid outbreak response for identification, confirmation and containment of possible epidemic/pandemic influenza and providing valid inclusion guidance for annual influenza vaccine formulation.
ACKNOWLEDGEMENTS
This work was funded in part by grants from the Influenza Division of the U.S. CDC and the U.S. Department of Defenses' Global Emerging Infection Systems (DoD-GEIS). The authors are grateful to the clinicians and staff at the field sites in Cambodia for their assistance in enrolling and sampling patients. We thank the laboratories at NAMRU2-Phnom Penh and the CDC for the conduct of the work. The views expressed herein are those of the authors and do not represent those of the U.S. Department of Health and Human Services, Department of Defense or Department of the Navy.
DECLARATION OF INTEREST
None.








