Crimean-Congo haemorrhagic fever (CCHF) is a fatal viral haemorrhagic fever with an overall fatality of 9–50% [Reference Ergonul1]. It is a tick-borne disease caused by the CCHF virus (CCHFV), a member of the genus Nairovirus, family Bunyaviridae. Humans are infected with CCHFV by the bite of infected Ixodid ticks, particularly ticks of the genus Hyalomma or contact with blood or tissue from viraemic livestock or human patients [Reference Hoogstraal2, Reference Rodriguez3]. The incubation period ranges from 2 to 9 days depending upon the route of exposure and viral inoculum [Reference Papa4]. The spread of CCHF disease may depend upon many factors such as trade and exchange of livestock, transmission via migratory birds infested with infected ticks and subsequent infection of new hosts and transmission in noscocomial settings due to direct contact [Reference Hoogstraal2, Reference Rodriguez3, Reference Tonbak5, Reference Burney6].
The CCHFV genome is triple-segmented having a length of ~19·2 kb. It has negative-sense, single-stranded RNA with segments, i.e. small (S), medium (M), and large (L). The L segment is ~12 kb long and encodes for non-structural proteins which comprise the RNA-dependent RNA polymerase gene. The M (length ~5·7 kb) and S (length ~1·7 kb) segments encode for structural proteins for the surface glycoproteins and nucleocapsid, respectively [Reference Zhou7, Reference Hewson8]. The CCHFV strains are clustered into seven genetically distinct groups on the basis of the S segment, i.e. Africa 1 (West Africa, Senegal), Africa 2 (Central, Democratic Republic of the Congo, South Africa), Africa 3 (South and West Africa), Europe 1, Europe 2 (Greece), Asia 1 (Middle East, Pakistan, Iran) and Asia 2 (China, Tajikistan, Kazakhstan) [Reference Carroll9–Reference Chamberlain12].
The geographical extent of CCHFV is wide-ranging, which can be correlated by the global distribution of the tick vector. The presence of CCHFV has been reported from many countries of Asia, Africa, South-East Europe and the Middle East. Frequent outbreaks of CCHF have been reported from countries bordering India such as Pakistan, Afghanistan, and western China. CCHF viral infection was reported in humans for the first time in Ahmadabad, Gujarat during 2011, although previous seroprevalence studies have shown viral antibodies both in animals and humans [Reference Yadav13]. The present study was undertaken for molecular characterization of CCHFV in India using the partial nucleocapsid (N) gene of the S segment.
Serum samples from three clinical cases of haemorrhagic fever, referred to National Centre for Disease Control (NCDC) from different geographical locations in India (Rajasthan and Uttar Pradesh), were tested for CCHFV (Fig. 1). Patients had constitutional symptoms, e.g. fever, arthralgia, and haemorrhagic manifestations, e.g. haematemesis, malena, epistaxis, etc. (Table 1). These samples belonged to focal outbreaks, therefore no ethical clearance was required.

Fig. 1. Map of India. Shaded areas indicate the location of the present study.
Table 1. Details of studied Crimean-Congo haemorrhagic fever virus patients
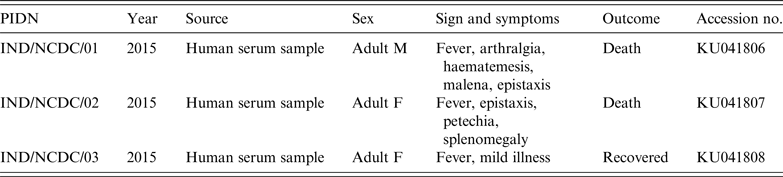
PIND, Patient identification number; M, male, F, female.
Viral RNA was isolated from the serum samples using QIAamp Viral RNA Mini kit (Qiagen, Germany) according to manufacturer's instructions. The RNA was finally eluted in 50 µl elution buffer provided in the kit and stored at −70 °C until use. Reverse transcription–polymerase chain reaction (RT–PCR) was performed using SuperScript One-step RT–PCR kit (Invitrogen Corp., USA). The outer PCR of 530 bp of the nucleocapsid (N) gene of CCHFV was amplified with forward primer: (CCHF-F2) TGGACA CCT TCA CAA ACT C, and reverse primer: (CCHF-R3) GAC AAA TTC CCT GCA CCA. The nested PCR of 226 bp was performed using forward primer: (CCHF-F3) GAA TGT GCA TGG GTT AGC TC, and reverse primer: (CCHF-R2) GAC ATC ACA ATT TCA CCA GG [Reference Drosten14]. Amplified products were visualized on 2% agarose gel with 0·5 µg/ml ethidium bromide. The PCR products were purified using the QIAquick PCR purification kit according to the manufacturer's instructions (Qiagen). Automated nucleotide sequencing was performed using the Big Dye Terminator Cycle Sequencing ready reaction kit (Applied Biosystems, USA) on an ABI 3130xl automated capillary DNA sequencer (Applied Biosystems). The nested PCR primers, CCHF-F3 and CCHF-R2, were used as forward and reverse primers, respectively, for sequencing. The sequences were aligned, edited and analysed using Clustal W multiple alignment. A BLAST (Basic Local Alignment Search Tool) search was carried out to compare data with already available sequences in the National Centre for Biotechnology Information (NCBI) database. The sequences were submitted to GenBank. Phylogenetic analysis was performed using molecular evolutionary genetics analysis (MEGA) software version 6.0 (http://www.megasoftware.net/) and a phylogenetic tree was generated using the neighbour-joining (NJ) method with bootstrap analysis of 1000 replicates using study sequences and other CCHF strains, which were retrieved from GenBank.
In the present study, all three studied serum samples from north-western India were found positive by RT–PCR of the N gene region of CCHFV, which were further sequenced. The sequences were submitted to GenBank and accession numbers were obtained (KU041806–KU041808). The sequences from the study shared 100% nucleotide identity among themselves. The sequences of the present study shared almost 99–100% nucleotide identity with strains from neighbouring countries like Pakistan, Afghanistan and Iran. The deduced amino-acid sequences from the study indicated no insertion/deletion in any of the analysed sequences. The study sequences shared a mean 94·04% nucleotide identity with earlier outbreak strains (2010–2011); reported from the western part of India.
The phylogenetic tree revealed that all CCHFVs divide in to seven groups, i.e. Asia 1, Asia 2, Africa 3, Africa 2, Europe 1, Africa 1 and Europe 2 (Fig. 2). The studied CCHFV sequences, clustered with strains previously reported from Pakistan, Afghanistan and Iran from different years, placed in the Asia 1 group. The CCHFV strains from UAE, Oman, Iraq and Madagascar formed a different cluster within the Asia 1 group. However, earlier (2010–11) Indian CCHFV isolates along with a large number of isolates from Tajikistan, Kazakhstan and Uzbekistan formed a separate cluster of the Asia 2 group.

Fig. 2. Phylogenetic tree of Crimean-Congo haemorrhagic fever virus (CCHFV) based on 220-bp nucleotide sequences of the N gene region generated by the neighbour-joining method. Bootstrap support values (based on 1000 replications) above 50% are shown at the branch nodes. Naming scheme is accession number followed by name of country. CCHFV strains that were sequenced in the study are shown in bold and indicated by a triangle (▲). Bold and italic indicate virus sequences previously reported from India.
Our findings reveal an emerging diversity of CCHFV strains circulating in India. The current CCHFV sequences belong to the Asia 1 group and previously reported CCHFV strains belong to the Asia 2 group [Reference Yadav13]. This study provides information on gene heterogeneity, phylogenetic relatedness and changing molecular epidemiology of the virus in India. Further studies are necessary to evaluate the impact of the emergence of diverse strains; their clinical manifestation and virulence.
ACKNOWLEDGEMENTS
The authors are grateful for the financial support received from the National Centre for Disease Control (NCDC) during the course of this study.
DECLARATION OF INTEREST
None






