Branched chain amino acids, particularly leucine, are the proteogenic amino acids involved in the regulation of animal physiology( Reference Wolfson, Chantranupong and Saxton 1 ). In addition to serving as a substrate for protein synthesis, leucine is also involved in several cellular processes, such as protein synthesis, energy metabolism and tissue regeneration. Several studies have indicated that leucine can regulate the mitochondrial biogenesis of skeletal muscle( Reference Sun and Zemel 2 , Reference D’Antona, Ragni and Cardile 3 ), glucose uptake( Reference Nishitani, Matsumura and Fujitani 4 – Reference Chang and Goldberg 6 ) and fatty acid oxidation( Reference Paul and Adibi 7 , Reference Li, Xu and Lee 8 ).
The skeletal muscle is an adaptive tissue involved in energy metabolism and protein turnover, and the main fibre types can be determined by the myosin heavy chain (MyHC) isoform expressed, namely, slow-oxidative type I (MyHC I), fast-oxidative type IIa (MyHC IIa), intermediate type IIx (MyHC IIx) and fast-glycolytic type IIb (MyHC IIb)( Reference Pette and Staron 9 , Reference Schiaffino and Reggiani 10 ). Most muscles in mammals contain a mixture of fibre types, and the fibre phenotype demonstrates high plasticity because of the diverse components of muscle fibres that are susceptible to the changes in nutrition, physical activity and environmental conditions( Reference Matsakas and Patel 11 , Reference Davidsen, Gallagher and Hartman 12 ). Furthermore, skeletal muscles are of critical importance as storage organs for energy and as materials for maintenance of the immune system and many metabolic pathways( Reference Ohlendieck 13 , Reference Reeds, Fjeld and Jahoor 14 ) under the condition of weaning stress.
Previous studies have shown that leucine stimulates protein synthesis in skeletal muscles through mechanistic targeting of the rapamycin (mTOR) pathway in rats( Reference Anthony, Anthony and Kimball 15 – Reference Mobley, Fox and Thompson 17 ), neonatal pigs( Reference Escobar, Frank and Suryawan 18 – Reference Hernandez-Garcia, Columbus and Manjarin 20 ) and humans( Reference Koopman, Verdijk and Manders 21 – Reference Kramer, Verdijk and Hamer 23 ), whereas the increased rate of protein synthesis caused by high-leucine levels does not necessarily lead to lean mass or body mass accretion. Moreover, several studies have shown that undernutrition decreases fibre size( Reference Stickland, Widdowson and Goldspink 24 ) and has more an important impact on type II fibres than on other types( Reference Goldspink and Ward 25 , Reference Sieck, Lewis and Blanco 26 ). In addition, Xia et al.( Reference Xia, Cholewa and Zhao 27 ) reported dietary supplementation with leucine combined with moderate aerobic training effectively increases fast-twitch muscle fibres. However, the proteome profile alterations caused by dietary leucine in the skeletal muscle of piglets remains elusive.
Proteomic analysis has been used to determine the markers of fibre-type specification( Reference Okumura, Hashida-Okumura and Kita 28 – Reference Gelfi, Vigano and De Palma 30 ), the metabolic processes in muscle development( Reference Gonnet, Bouazza and Millot 31 , Reference Chan, McDermott and Siu 32 ), the effect of exercise, weight loss( Reference Guelfi, Casey and Giles 33 , Reference Almeida, van Harten and Campos 34 ) and the pathological effects of type 2 diabetes and ageing-related changes on skeletal muscle( Reference Hwang, Bowen and Lefort 35 – Reference Gannon, Staunton and O’Connell 37 ). In addition, Wang et al.( Reference Wang, Ou and Yin 38 ) reported that ZnO supplementation improves the redox state and prevents apoptosis in the jejunum of weanling piglets using two-dimensional gel electrophoresis and MS. Goichon et al.( Reference Goichon, Chan and Lecleire 39 ) revealed that leucine supplementation may slow down fatty acid β-oxidation (FAO) in human duodenal mucosa using the same methods. Thus, this study was designed to investigate the alterations of differentially expressed proteins and their metabolic pathways, regulated by leucine, in the skeletal muscle of weanling piglets.
Methods
Animals and diets
All procedures on animals and all the experiments were conducted in accordance with the ethical guidelines for animal research established and approved by the Institutional Animal Care and Use Committee at Huazhong Agricultural University. A total of forty-eight male piglets (Yorkshire×Large White, weaned at 21 d of age) were housed in six pens of eight piglets each, with the pens having slotted stainless steel floors, and assigned randomly to one of two dietary groups (three pens per group). The control group was fed the basal diet containing 1·66 % leucine (normal leucine level (NL)). The treatment group was fed the basal diet supplemented with leucine to provide 2·1 % leucine (high-leucine diet (HL)). The temperature was kept at 25–28°C day–night. Each pen was equipped with a feeder and a nipple waterer to allow all piglets free access to feed and water. Growth performance of the weanling piglets in the NL and the HL groups are shown in the online Supplementary Table S1. One piglet per pen was slaughtered to collect longissimus dorsi (LD) muscle samples at 35 d of age after weaning. Muscle samples were placed in liquid N2 and stored at –80°C.
Reagents
The following antibodies were used: antibodies to medium-chain-specific acyl-CoA dehydrogenase (ACADM) (A1873), ATP synthase subunit b (ATP5F1) (A7645), glutathione S-transferase pi 1(GSTP1) (A5691), long-chain 3-ketoacyl-CoA thiolase (HADHB) (A5716), dual specificity mitogen-activated protein kinase kinase 6 (MAP2K6) (A2575), MyHC IIx (A6935), MyHC IIa (A3620), MyHC I (A7564), 40S ribosomal protein S4, X isoform (RPS4X) (A6730), troponin I protein (TNNI) 1 (A4161), TNNI2 (A7937) and α-tubulin (AC007) from ABclonal; antibodies to AMP-activated protein kinase (AMPK) (sc-74461) and p-AMPK (Thr172, sc-33524) from Santa Cruz Biotechnology; antibodies to myostatin (MSTN) (D160418), PPARγ coactivator-1α (PGC-1α) (D162041), pyruvate kinase (PKM) (D120008) and succinate dehydrogenase (SDH) B (D162175) from BBI Life Sciences; antibodies to mTOR (2983), p-mTOR (Ser2448, 2971), p70S6K (2708) and p-p70S6K (Thr389, 9234) from Cell Signaling Technology; antibodies to MyHC IIb (20140-1-AP), PGC-1β (22378-1-AP) from Protein Tech Group; HRP-conjugated anti-mouse secondary antibody from Cell Signaling Technology; and anti-rabbit secondary antibody from Sigma.
Protein sample preparation, digestion and isobaric tags for relative and absolute quantitation labelling
Muscle samples were ground into a powder in liquid N2; 150 mg powder was extracted with 1 ml lysis buffer (7 m urea, 2 m thiourea, 4 % 3-[(3-cholamido-propyl)-dimethylammonio]-1-propanesulfonate (CHAPS), 40 mm TRIS-HCl, pH 8·5, 10 mm dithiothreitol containing 1 % protease inhibitor cocktail (P8340; Sigma) on ice surface, fully homogenised and then centrifuged at 4°C, 12 000 g for 15 min. The supernatants were collected to new clean tips and the protein concentration was quantified using the Bradford method. To reduce the effect of variation in biological samples on the proteomic analysis, three muscle samples from each group were pooled together and stored at –80°C. These mixed protein samples were used for isobaric tags for relative and absolute quantitation (iTRAQ) and Western blotting assays.
A total of 100 mg mixed protein from each group was digested with Trypsin Gold (Promega) with the weight ratio of protein:trypsin=30:1 at 37°C for 16 h. Then, the digested peptides were vacuum centrifuged for drying, reconstituted in 0·5 m tetraethylammonium bromide (Applied Biosystems) and labelled according to the manufacture’s protocol for 8-plex iTRAQ reagent (Applied Biosystems) as follows: the NL sample was labelled with 119-tag and 121-tag. The peptides were labelled with the isobaric tags and incubated at room temperature for 2 h. The labelled peptide were then pooled and dried using vacuum centrifugation.
Strong Cation Exchange fractionation and liquid chromatography--electrospray ionisation-MS/MS analysis
Labelled peptides were separated by Strong Cation Exchange (SCX) chromatography using LC-20AB HPLC Pump system (Shimadzu). The dried peptide mixture was reconstituted with 4 ml buffer A (25 mm NaH2PO4 in 25 % acetonitrile (ACN), pH 2·7) and gradient eluted with buffer B (25 mm NaH2PO4, 1 m KCl in 25 % ACN, pH 2·7). Peptides were loaded onto a 4·6×250 mm Ultremex SCX column (Phenomenex) with the following elution procedure: flow rate, 1 ml/min; buffer A for 10 min; buffer B, 5–60 % for 27 min, 60–100 % for 1 min. Elution was monitored by measuring the absorbance at 214 nm. A total of twenty fractions were collected and desalted with a Strata X C18 column (Phenomenex).
Each fraction was dried and resuspended in buffer A (2 % ACN, 0·1 % formic acid) for LC-MS/MS. On average, the final concentration of peptides was about 0·5 μg/μl, following which 10 μl of each fraction was loaded on a LC-20AD nanoHPLC (Shimadzu) using a 2-cm C18 trap column. Later, the peptides were eluted onto a 10-cm analytical C18 column (inner diameter 75 μm) using the following procedure: loading 8 μl/min for 4 min, then subjecting to a gradient of 2–35 % in buffer B (98 % ACN, 0·1 % formic acid) at 300 nl/min for 44 min, followed by 2 min linear gradient to 80 % and maintenance at 80 % in buffer B for 4 min, and finally returning to 5 % in 1 min. Next, all peptides were identified using tandem MS (MS/MS) in QEXACTIVE (Thermo Fisher Scientific) coupled online to the HPLC. Intact peptides were detected using Orbitrap (Thermo Fisher Scientific) at a resolution of 70 000 and MS scans were obtained from 350 to 2000 m/z. A total of fifteen precursor ions were selected using high-energy collision dissociation with a dynamic exclusion duration of 15 s. The MS/MS scan range was 100–1800 m/z.
Data analysis
Protein identification was performed using Mascot search engine (version 2.3.02, Matrix Science) embedded into Proteome Discoverer 1.2 against the Uniport database containing pig protein sequences. For protein identification, a mass tolerance of 20 parts per million was permitted for intact peptide masses and 0·1 Da was permitted for fragmented ions. To minimise false-positive results, we counted only peptides at the 95 % CI and a false discovery rate ≤0·01, containing at least one unique peptide. For protein quantitation, a differentially expressed protein containing at least two unique spectra was used with a fold-change value >1·2 or <0·83 (1/1·2) with P<0·05 between the NL and the HL groups. Protein subcellular localisation prediction was on the basis of Gene Ontology (GO) annotation. The corresponding pathways that the proteins mapped were searched using online Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The functional classification, network analysis and pathway analysis of differentially expressed proteins were analysed by core analysis using Ingenuity Pathway Analysis (IPA) software.
Western blotting analysis
Tissue lysates were mixed with the 5× protein loading buffer (161-0737; Bio-Rad). After heat denaturation, equal amounts (15 μg) of cellular proteins were separated using SDS-PAGE, followed by electrotransfer onto a polyvinylidene fluoride (PVDF) membrane (162-0177; Bio-Rad). After blocking with 5 % non-fat milk, the PVDF membrane was performed by incubation with primary antibodies (1:2000 dilution), then with secondary antibodies (1:10 000 dilution), washing after each step with TBST (Tris-buffered saline, 0·1 % Tween 20) three times for 5 min. Washing was carried out with TBS two times for 5 min before detection with Immobilon Western Chemiluminescent HRP substrate (no. WBKLS0500; Millipore).
ATP and AMP assay
The ATP and AMP levels in the skeletal muscle were determined by HPLC analysis. A quantity of 100 mg muscle powder was extracted with 1 ml 7 % (w/v) HClO4 on an ice surface. After centrifugation at 15 000 g for 10 min at 4°C, the supernatant was removed and mixed with an equal volume of 1·03 m KOH, which was centrifuged again at 15 000 g for 10 min at 4°C; 20 μl of supernatant was loaded on a LC-20AT HPLC (Shimadzu) by an analytical column (250mm×4·6mm, internal diameter, 5 μm; Hypersil ODS2; Elite). The UV detector is set at 259 nm.
Statistical analysis
All statistical analyses were carried out with GraphPad Prism 5.01.336 (GraphPad Software). The paired t test was used for statistical analysis between two groups. The measurements were expressed as means with their standard errors. The significance level was set at P<0·05; * P<0·05; ** P<0·01; *** P<0·001.
Results
Quantitative proteomic analysis of skeletal muscle after leucine supplementation
To assess the differences in skeletal muscle proteome altered by leucine level, we analysed the LD muscle samples derived from the NL and the HL groups in which iTRAQ tags were applied (NL: 119 and HL: 121) (Fig. 1(a)). In this case, we showed that 157 quantifiable proteins were significantly altered by leucine supplementation, containing sixty-one up-regulated (fold change >1·2 with P<0·05) and ninety-six down-regulated (fold change <0·83 with P<0·05) proteins (Fig. 1(b) and online Supplementary Table S2). Heat map in different colours was used to show the differentially expressed protein profiles between the NL and the HL groups, and all the differentially expressed proteins are shown in Fig. 1(c).

Fig. 1 Schematic workflow of isobaric tags for relative and absolute quantitation (iTRAQ) and differentially expressed proteins are identified between high-leucine diet (HL)/normal leucine diet (NL) groups. (a) Experimental design and workflow of this study. Longissimus dorsi muscle tissue samples were individually collected from two groups feeding with different level of leucine (1·66 %, NL or 2·1 %, HL) in diet at 35 d of age after weaning. Three muscle samples from each group were pooled together. A total of two pooled samples per treatment were used for MS analysis and labelled with different iTRAQ tags (iTRAQ-119 or iTRAQ-121). (b) The total number of up-regulated and down-regulated proteins. (c) Heat map showed the 157 differentially expressed proteins identified and quantified. Differentially expressed proteins are defined by the iTRAQ ratio: up-regulated proteins, HL:NL >1·2, P<0·05; down-regulated proteins, HL:NL <0·833, P<0·05.
Functional classification, subcellular localisation and network analysis for differentially expressed proteins
To unambiguously analyse the molecular functions of each of these differentially expressed proteins, we used the IPA software to get the functional classification. Fig. 2(a–c) shows the distribution charts of diseases and disorders, molecular and cellular function, and the physiological system development and function. (A complete catalogue of each part is exhibited in the online Supplementary Table S3). We predicted that these significantly altered proteins may be involved in enhanced processes, such as skeletal and muscular disorders (Fig. 2(a)), energy production, lipid metabolism and protein synthesis (Fig. 2(b)), cell morphology, organ morphology and skeletal and muscular system development and function (Fig. 2(c)).

Fig. 2 Functional analysis, subcellular localisation and key network analysis for differentially expressed proteins in high-leucine diet group compared with normal leucine diet group. The items of diseases and disorders (a), molecular and cellular functions (b) and physiological system development and function (c) categories were listed by P value (![]() , –log P). (d) Subcellular localisation of the up-regulated proteins (
, –log P). (d) Subcellular localisation of the up-regulated proteins (![]() ) and down-regulated proteins (
) and down-regulated proteins (![]() ) for differentially expressed protein. (e) The top diseases and function networks about the differentially expressed proteins caused by leucine supplementation.
) for differentially expressed protein. (e) The top diseases and function networks about the differentially expressed proteins caused by leucine supplementation.
To assess the organelle specificity of the differentially expressed proteins, we analysed the subcellular localisation according to their GO annotation. All categories were ranked by the numbers of differentially expressed proteins. These results showed that the cytoplasm, mitochondrion, membrane and the nucleus were enriched with both up-regulated proteins and down-regulated proteins (Fig. 2(d)).
In the next step, we analysed the interaction network of the 157 differentially expressed proteins (online Supplementary Table S4). Proteins were enriched in two networks, predominantly, ‘energy production, lipid metabolism, small molecule biochemistry’ and ‘protein synthesis, energy production, nucleic acid metabolism’ (Fig. 2(e)).
Leucine supplementation alters energy metabolism in muscle tissue
To gain a biological insight into differentially expressed proteins, we analysed the canonical pathways using IPA (online Supplementary Table S5). The top canonical pathways were ranked by P value (Fig. 3(a)) and the proteins enriched in each pathway were presented (Fig. 3(b)). The study results indicated that many energy production pathways were enriched. The oxidative phosphorylation (OXPHOS) pathway showed a minimum P value (Fig. 3(a)) and a total of fourteen proteins (ATP synthase subunit delta (ATP5D), ATP synthase subunit b (ATP5F1), ATP synthase subunit e (ATP5I), ATP synthase-coupling factor 6 (ATP5J), cytochrome c oxidase subunit 5A (COX5A), cytochrome c oxidase subunit 5B (COX5B), cytochrome c oxidase subunit 6C (COX6C), cytochrome c oxidase subunit 7C (COX7C), cytochrome c oxidase subunit 2 (MT-CO2), NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 7 (NDUFA7), NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 9 (NDUFB9), succinate dehydrogenase [ubiquinone] iron-sulfur subunit (SDHB), succinate dehydrogenase cytochrome b560 subunit (SDHC), succinate dehydrogenase [ubiquinone] cytochrome b small subunit (SDHD)) were identified as the members of this pathway. Notably, twelve of fourteen identified proteins were down-regulated in the HL group (Table 1), which suggests that the OXPHOS pathway is suppressed by leucine supplementation. We further detected SDHB (0·761-fold in iTRAQ) and ATP5F1 (0·763-fold), the enzymes that participated in the OXPHOS pathway( Reference Oyedotun and Lemire 40 ), which were significantly decreased in the HL group (P<0·05) (Fig. 3(c) and (d)). However, the study data also showed an increase in the expression of PKM (1·269-fold) in the glycolysis pathway in the iTRAQ experiment (Table 1) and immunoblotting analysis in the HL group (Fig. 3(e)). However, three of five quantified proteins in the FAO pathway were decreased in response to leucine supplementation (Table 1). HADHB (0·754-fold) and ACADM (0·759-fold) are two key proteins in FAO, and showed a decline in the HL group (Fig. 3(f) and (g)). Moreover, we showed that ATP concentration was significantly decreased in the leucine-supplemented group (P<0·05) (Table 2). Overall, these results suggest that dietary supplementation with leucine suppressed OXPHOS and FAO pathways, and up-regulated the glycolysis pathway in the LD muscle of weanling piglets.

Fig. 3 Oxidative phosphorylation pathway and fatty acid β-oxidation pathway are suppressed, and glycolysis pathway is activated by leucine in skeletal muscle of weanling piglets. Top pathways (a) and significantly regulated proteins were clustered (b) (![]() , up-regulated proteins, 1·2-fold;
, up-regulated proteins, 1·2-fold; ![]() , down-regulated proteins, –1·2-fold) in each pathway in response to leucine. The expression of SDHB (c), ATP5F1 (d), PKM (e) and ACADM (f), HADHB (g) in two groups were detected by Western blotting analysis. Values are means (n 5 experiments) with their standard errors represented by vertical bars. * P<0·05; ** P<0·01. α-Tubulin was detected as a loading control. HL, high-leucine diet; NL, normal leucine diet.
, down-regulated proteins, –1·2-fold) in each pathway in response to leucine. The expression of SDHB (c), ATP5F1 (d), PKM (e) and ACADM (f), HADHB (g) in two groups were detected by Western blotting analysis. Values are means (n 5 experiments) with their standard errors represented by vertical bars. * P<0·05; ** P<0·01. α-Tubulin was detected as a loading control. HL, high-leucine diet; NL, normal leucine diet.
Table 1 List of candidate proteins and pathways in response to leucine supplementation

HL, high-leucine diet; NL, normal leucine diet; EIF2, eukaryotic initiation factor 2; NRF2, nuclear factor-erythroid 2-related factor 2; mTOR, mechanistic target of rapamycin.
* P<0·05.
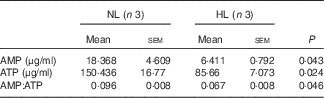
Table 2 The concentration of AMP and ATP in longissimus dorsi muscle from normal leucine diet (NL) and high-leucine diet (HL) groups (Mean values with their standard errors)
Leucine supplementation promotes protein synthesis and alters the constitution of muscle fibres
The iTRAQ data showed a low P value for the eukaryotic initiation factor 2 (EIF2) and the mTOR signalling pathways (Fig. 4(a) and online Supplementary Table S5). Among the nine identified differentially expressed proteins in EIF2 signalling, seven proteins (eukaryotic translation initiation factor 4 gamma 1 (EIF4G1), 60S ribosomal protein L36 (RPL36), 60S ribosomal protein L27A (RPL27A), 40S ribosomal protein S30 (FAU), 40S ribosomal protein S15A (RPS15A), 40S ribosomal protein S27A (RPS27A), 40S ribosomal protein S4, X isoform (RPS4X)) were significantly up-regulated in the HL group (Table 1). We observed that the expression of RPS4X and the ratio of p-mTOR:mTOR and p-p70S6K:p70S6K were increased in the HL group (Fig. 4(a–c)). These results indicated that leucine supplementation promoted protein synthesis in the skeletal muscle tissue.

Fig. 4 Leucine supplementation promotes protein synthesis and slow-to-fast muscle fibre transitions. The expression of p-mTOR, mTOR (a), p-p70S6K, p70S6K (b), RPS4X (c), TNNI1 (e), TNNI2 (f), MyHC I (g), MyHC IIa (h), MyHC IIx (i) and MyHC IIb (j) induced by leucine were confirmed by Western blotting analysis. (d) Myofibrillar proteins identified in high-leucine diet (HL) group compared with normal leucine diet (NL) group. Values are means (n 4 experiments) with their standard errors represented by vertical bars. * P<0·05; ** P<0·01; *** P<0·001. α-Tubulin was detected as a loading control. HL, high-leucine diet; NL, normal leucine diet.
Furthermore, we identified eight myofibrillar proteins in the iTRAQ data (Fig. 4(d)). These are mainly due to the differences in the muscle fibre types. TNNI1 and TNNI2 have been used as model genes to study the mechanisms of slow and fast fibre-specific expression. The data from the present study show that these two proteins have the ratio changed between HL and NL but with no significance. We tested the decreased expression of TNNI1 and increased expression of TNNI2 by Western blotting analysis after leucine supplementation (Fig. 4(e) and (f)). We also detected the content of four myosin proteins, MyHC I, MyHC IIa and MyHC IIx were significantly decreased, whereas MyHC IIb was increased in the HL group (Fig. 4(g–j)). All these results reflect a possible modulation of slow-to-fast transitions.
Leucine supplementation decreases the activity of AMP-activated protein kinase and suppresses the p38/c-JUN NH2-terminal protein kinase pathway
We next examined PGC-1α and PGC-1β, the two key regulators for muscle fibre transitions, to explore how leucine is involved in muscle fibre transitions. PGC-1α showed no significant change (Fig. 5(a)), but PGC-1β was down-regulated in the HL group (Fig. 5(b)). AMPK, as the main sensor of intracellular energy status, is activated by an increase in the AMP:ATP ratio( Reference Hardie, Ross and Hawley 41 ). In the assay, the ratio of AMP:ATP and p-AMPK:AMPK are all decreased in the HL group (Fig. 5(c) and Table 2). These results suggest the inhibition of the activation of AMPK in leucine-supplemented piglets.

Fig. 5 Leucine supplementation decreases the activity of AMP-activated protein kinase (AMPK) and suppresses the p38/c-JUN NH2-terminal protein kinase pathway. Western blotting detected the expression of PPARγ coactivator-1α (PGC-1α) (a), PPARγ coactivator-1β (PGC-1β) (b), p-AMPK, AMPK (c), MAP2K6 (d), Glutathione S-transferase P1 (GSTP1) (e) and myostatin (MSTN) (f) in high-leucine diet (HL) group compared with normal leucine diet (NL) group. Values are means (n 5 experiments) with their standard errors represented by vertical bars. * P<0·05, **P<0·01. α-Tubulin was detected as a loading control.
The study data also identified MAP2K6 (also named as MKK6, 0·646-fold) and GSTP1 (1·443-fold) with significant changes in the HL group in the iTRAQ data (Table 1) and Western blotting analysis (Fig. 5(d) and (e)). MAP2K6 is one of major regulators for p38 mitogen-activated protein kinase (also named as p38) activation( Reference Deacon and Blank 42 , Reference Enslen, Raingeaud and Davis 43 ), and GSTP1 is considered to be an inhibitor for c-JUN NH2-terminal protein kinase (JNK) activation( Reference Hayes and Pulford 44 ). These results suggest the inhibition of p38/JNK pathway. In addition, MSTN is a member of the transforming growth factor-β family that is widely distributed in muscle tissue and is regarded as a negative regulator of muscle growth( Reference McPherron, Lawler and Lee 45 ). We detected that the expression of MSTN is declined in LD muscle derived from the HL group (Fig. 5(f)).
Discussion
A proteome-wide analysis on weanling piglets expands our knowledge of how leucine supplementation affects skeletal muscle metabolism. To our knowledge, this is the first report of proteomic analysis that analysed the effects of dietary supplementation with leucine in weanling piglets. In the present study, the 157 differentially expressed proteins between the NL and the HL groups, altered by dietary leucine supplementation, demonstrate the changes in energy metabolism, protein synthesis and fibre-type distribution in the LD muscle of weanling piglets.
Muscles, which convert chemical energy into mechanical movement, are of critical importance as storage organs for energy, as well as for the maintenance of energy homoeostasis. Two core metabolic pathways, glycolysis and OXPHOS, are involved in ATP production for energy, whereas glycolysis is a catabolic process that converts glucose to pyruvate, which then can be completely oxidised to produce maximal amounts of ATP through the OXPHOS pathway. Besides glucose, fatty acid and amino acid also serve as energy substrates involved in mitochondrial oxidation( Reference Newgard 46 ). Indeed, in skeletal muscle, the regulation of energy homoeostasis in different conditions triggers whole rearrangement of glucose and lipid metabolisms( Reference Gerhart-Hines, Rodgers and Bare 47 ), which always is accompanied by the modulation of glycolysis or OXPHOS. In this case, we identified that fourteen differentially expressed proteins in OXPHOS pathway responded to leucine supplementation, which suggests that high-level leucine suppresses the OXPHOS pathway. With this in perspective, we showed a suppressed FAO and a lower ATP content in the HL group. Moreover, compared with the NL group, the study results in the iTRAQ experiment exhibited an increased abundance of hexokinase 1 (1·246-fold) and PKM (1·269-fold) in leucine-supplemented piglets, which results in enhanced glycolytic activity. Thus, high-level leucine leads to a shift from oxidative to glycolytic metabolism in the LD muscle of weanling piglets.
Because of the alterations in fibre type distribution, skeletal muscle fibres exhibit a physiological plasticity adapting to the diverse conditions( Reference Fluck and Hoppeler 48 , Reference Spangenburg and Booth 49 ). Of the two main fibre types in skeletal muscle, the fast type IIb fibre has less mitochondrion but higher activity of glycolytic enzymes than the other type. Lombardi et al.( Reference Lombardi, Silvestri and Cioffi 50 ) reported an increase in various oxidative enzymes and a decrease in glycolytic enzymes in aged gastrocnemius of rats with the increase in slow-type myosin proteins and the decrease in fast-type myosin proteins. They concluded a transition from fast-to-slow muscle fibre and a glycolytic-to-oxidative metabolic shift during skeletal muscle ageing. Moreover, chronic stimulation of β2-adrenoceptors induces shifts from the MyHC I to the MyHC II isoform and from an oxidative to a glycolytic phenotype in rats( Reference Ohnuki, Umeki and Cai 51 ). These results indicate that the alterations in energy metabolic pathways significantly correlate with muscle fibre transitions.
Many myofibrillar proteins have been shown in the study data. Troponin T protein (TNNT), fast skeletal muscle (TNNT3, 2·126-fold) and troponin I, fast skeletal muscle (TNNI2, 1·253-fold, non-significant in iTRAQ), the specific proteins in fast muscle fibres( Reference Muroya, Nakajima and Chikuni 52 ), all show an increased expression in the HL group. Whereas, troponin I, slow skeletal muscle (TNNI1, 0·68-fold, non-significant in iTRAQ) and tropomyosin α-3 chain (0·75-fold), which are mainly expressed in slow muscle fibres( Reference Perry 53 ), are all down-regulated by leucine supplementation. Furthermore, the fast fibre types show the lowest activity of SDH( Reference Burke, Levine and Zajac 54 ). The study results show that the expression of three enzymes SDHB (0·761-fold), SDHC (0·793-fold) and SDHD (0·741-fold) are decreased in the HL group. We also detected a decreased expression of MyHC I, MyHC IIa, MyHC IIx and an increased expression of MyHC IIb in the HL group. All these results suggest that leucine supplementation increases the growth of fast-twitch muscle fibres and decrease in the slow type. Thus, the up-regulated glycolysis and down-regulated OXPHOS in the LD muscle explains that, probably, leucine supplementation yields a higher percentage of glycolytic type II fibres during postweaning time.
Previous studies have corroborated that leucine alone is sufficient to regulate the activity of the mTOR pathway( Reference Hara, Yonezawa and Weng 55 ) and that it acts as a modulator of muscle protein synthesis( Reference Patti, Brambilla and Luzi 56 , Reference Anthony, Anthony and Kimball 57 ). In contrast, as a negative regulator for muscle growth, MSTN can inhibit the mTOR-mediated protein synthesis pathway( Reference Elliott, Renshaw and Getting 58 ) and activate the protein degradation pathway( Reference Mendias, Kayupov and Bradley 59 , Reference Lokireddy, McFarlane and Ge 60 ). Liu et al.( Reference Liu, Pan and Li 61 ) reported that maternal low-protein diet could up-regulate MSTN in the skeletal muscle of weanling piglets. They also reported a decreased leucine level in blood and inhibition of mTOR pathway in the skeletal muscle of weanling piglets. The study results also found that the ratios of p-mTOR:mTOR and p-p70S6K:p70S6K were increased in the HL group. In addition, EIF4G1 and proteins from 60S ribosomal subunit (RPL36 and RPL27A) and 40S ribosomal subunit (FAU, RPS15A, RPS27A and RPS4X) were all up-regulated in the leucine-supplemented condition. Moreover, the iTRAQ data also showed that the proteins in the protein ubiquitination pathway (beta-2-microglobulin (B2M), alpha-crystallin B chain (CRYAB), DnaJ homolog subfamily B member 4 (DNAJB4), heat shock 70 kDa protein 1A (HSPA1A), heat shock protein beta-2 (HSPB2), heat shock protein beta-6 (HSPB6), heat shock protein beta-7 (HSPB7), heat shock protein beta-8 (HSPB8), 26S protease regulatory subunit 7 (PSMC2), proteasome activator complex subunit 1 (PSME1)) were all decreased in the HL group (Table 1). These results suggest that dietary leucine supplementation promotes muscle protein synthesis, and that leucine might inhibit the function of MSTN in skeletal muscle.
Numerous studies have shown that loss of MSTN leads to an increased proportion of fast type II fibres and a reduced proportion of slow type I fibres in littermates or aged muscles( Reference Siriett, Platt and Salerno 62 , Reference Jackson, Luong and Vang 63 ). This phenomenon suggests that the loss of MSTN might cause a transformation to a more fast-twitch phenotype in skeletal muscle. Moreover, MSTN is a secreted protein from the skeletal muscle( Reference McPherron, Lawler and Lee 45 ), and two receptors named activin receptor type IIB and activin receptor-like kinase (ALK4/5) initiate MSTN signalling( Reference Lee and McPherron 64 , Reference Rebbapragada, Benchabane and Wrana 65 ). MSTN can inhibit AMPK through its receptors( Reference Biesemann, Mendler and Wietelmann 66 ), which was supported by the finding that MSTN-deficient mice exhibit the activation of the AMPK signalling pathway( Reference Zhang, McFarlane and Lokireddy 67 ). Interestingly, Das et al.( Reference Das, Yang and Fu 68 ) showed that AMPK can stimulate the expression of MSTN. These findings suggested that there might be a negative feedback loop between AMPK and MSTN. Thus, a possible explanation for the decreased MSTN expression and phosphorylation of AMPK in our results is that leucine supplementation inhibits the activation of AMPK, thus leading to the reduced expression of MSTN. The decreased MSTN may act to protect AMPK from over-inhibition by leucine. In addition, MSTN can induce the activation of either the p38( Reference Philip, Lu and Gao 69 ) or the JNK signalling pathway( Reference Huang, Chen and Zhang 70 ) in the adult murine myoblast cell line C2C12. However, Morissette et al.( Reference Morissette, Cook and Foo 71 ) observed that overexpression of MSTN did not increase the phosphorylation of p38 in neonatal rat cardiomyocytes. The decreased levels of MSTN and p38/JNK observed in the HL group implies that a high level of leucine in the diet promotes a more fast-twitch phenotype in the skeletal muscle of weanling piglets, and that MSTN might have a stage-dependent effect on p38.
PGC-1 has been proved to be a master regulator for cellular energy metabolism, and a key signalling molecule for fibre-type determination( Reference Lin, Handschin and Spiegelman 72 , Reference Finck and Kelly 73 ). Several reports demonstrated that PGC-1-mediated myofibre switching towards a slower and oxidative phenotype. Overexpression of PGC-1α in skeletal muscle leads to the increase of slow-type fibres and the expression of OXPHOS proteins( Reference Lin, Wu and Tarr 74 ). PGC-1β can drive the formation of oxidative type IIX fibres( Reference Arany, Lebrasseur and Morris 75 ). Although the upstream signalling pathways involved in the regulation of PGC-1 are still largely unknown, both AMPK and p38 can affect PGC-1α expression and activity( Reference Arany 76 ). As reported recently, AMPK has been associated with crucial roles in fibre-type transformation and mitochondrial biogenesis, mainly relying on the function of PGC-1α ( Reference Zong, Ren and Young 77 , Reference Roeckl, Hirshman and Brandauer 78 ). In addition, p38 has been shown to contribute to the induction of PGC-1α expression( Reference Akimoto, Pohnert and Li 79 ) and to directly phosphorylate PGC-1α ( Reference Fan, Rhee and St-Pierre 80 , Reference Puigserver, Rhee and Lin 81 ). However, the interaction between AMPK, p38 and PGC-1β expression is rarely reported. Simultaneously, PGC-1 have been shown to robustly induce the expression of a specific transcription factor myocyte enhancer factor-2( Reference Lin, Wu and Tarr 74 , Reference Arany, Lebrasseur and Morris 75 ), which can enhance the majority of muscle-specific gene expression( Reference McKinsey, Zhang and Olson 82 ), and is involved in the formation of slow-twitch muscle fibres( Reference Esser, Nelson and Lupa-Kimball 83 , Reference Chin, Olson and Richardson 84 ). In this case, our results indicated that PGC-1α showed no significant change, but PGC-1β was down-regulated, after leucine supplementation. We speculated that the decreased expression of MSTN and the inhibition of p38 and AMPK will lead to the decreased expression and activity of PGC-1β, but not of PGC-1α, which will promote the switching of slow muscle fibres towards the fast type after leucine supplementation.
In conclusion, we have performed a proteomic profiling to investigate the effects of leucine supplementation on the skeletal muscle proteome of weanling piglets. The results especially indicated that leucine supplementation may alter the energy metabolism and promote slow-to-fast transitions in the skeletal muscle of weanling piglets, which provide new insights into the regulation of energy homoeostasis as well as muscle fibre transitions through leucine. However, future studies are still needed to elucidate the underlying regulatory mechanism of specific amino acids, in particular, of leucine.
Acknowledgements
The authors gratefully acknowledge all present and past members of Yan laboratory who have contributed comments and ideas.
This work was supported by the National Natural Science Foundation of China (grant nos 31322053, 31520103915 and 31172290); the National Key Basic Research Program of China (973 Program) (grant no. 2013CB127305); the Hubei Province Distinguished Young Scholar (grant no. 2012FFA015) and the Fundamental Research Funds for the Central Universities (grant nos 2662015PY111 and 2013JQ001). No funding body had any role in the design, analysis or writing of this article.
Q. F. and X. Y. designed the study, interpreted the data and wrote the manuscript; Q. F. and B. L. performed the animal studies; Q. F., G. Y., M. S. and X. B. collected the data; Q. F., Z. W., J. H., X. L., C. C. and Z. Z. conducted the biochemical experiments and analysed the data. All authors contributed to the discussion and approved the final version of this paper.
The authors declare that there are no conflicts of interest.
Supplementary material
For supplementary material/s referred to in this article, please visit https://doi.org/10.1017/S0007114517001209









