Dengue is the most important arboviral infection of medical importance with around 40% of the global population now living in areas at risk. The WHO estimates that about 50 million dengue infections are occurring worldwide annually with ~25 000 deaths [1]. Dengue can be caused by any of the four antigenically distinct serotypes of dengue viruses (DENV-1 to -4). Dengue viruses belong to the genus Flavivirus of the family Flaviviridae. DENV are positive-sense single-stranded RNA viruses. The genome is ~11 kb in length [Reference Gubler2]. All the four DENV serotypes are primarily transmitted by Aedes (Stegomyia) species mosquitoes.
Recently, outbreaks of dengue infection have increased in many parts of the tropics and subtropics [1]. The incidence of dengue infections has also grown dramatically over the past two decades in many parts of India. The last decade witnessed dominance of different dengue serotypes in India. The largest dengue outbreak in India occurred in 1996 and the aetiology was identified as DENV-2 [Reference Dar3]. This was later replaced by DENV-3, as the dominant serotype in 2003 [Reference Dash4]. Subsequent outbreaks witnessed dominance of DENV-1 along with DENV-2 and -3 [Reference Dar5, Reference Kukreti6]. The involvement of DENV-4 is very rarely reported from isolated cases after the 1970s in India [Reference Dar5, 7]. Moreover, there is a lack in the available genetic information and genotyping of Indian DENV-4 viruses.
We described the investigation of a major dengue-like illness in Andhra Pradesh, India during October–December 2007. A total of 275 blood samples from patients suspected of having acute dengue fever were collected with informed consent from Nizam's Institute of Medical Sciences (NIMS), a tertiary-care research and referral Medical Institute in Hyderabad, Andhra Pradesh for this study. A set of blood samples was collected with and without anticoagulant for virus isolation and serology, respectively. All these samples were investigated for the presence of dengue-specific IgM and IgG antibodies by using IgM and IgG capture ELISA (PanBio, Australia). The presence of dengue-specific RNA in clinical samples was detected using the Access Quick One-Step RT–PCR kit (Promega, USA) employing a primer pair targeting the C-prM gene junction [Reference Lanciotti8, Reference Dash9]. The positive amplicons were subjected to nested PCR for serotyping [Reference Lanciotti8] and were also subjected to nucleotide sequencing employing the Big Dye Terminator Cycle Sequencing Ready Reaction kit with ABI 3100 sequencer (Applied Biosystems, USA) for confirmation. Further, complete envelope (E) gene of two DENV-4 isolates (ND73 and ND110) at first passage level were amplified and sequenced using specific primers as reported previously [Reference Klungthong10]. The sequences of envelope gene of ND73 and ND110 comprising 1485 nucleotides were submitted to GenBank under accession numbers HM237348 and HM237349, respectively. An extensive phylogenetic analysis based on complete E gene was performed by including 182 globally diverse DENV-4 E gene sequences, using MrBayes version 3.1.2 [Reference Ronquist and Huelsenbeck11]. The Bayesian tree was inferred by running Markov Chain Monte Carlo (MCMC) iterations for 1500 000 generations, sampling at every 100th generation with a burn-in setting of 10% generations. Convergence was assessed using average standard deviation in partition frequency values using a threshold of 0·01. The phylogenetic grouping was also confirmed using Neighbour-Joining (NJ) and Maximum-Parsimony (MP) methods using MEGA3 software version 3.1 [Reference Kumar, Tamura and Nei12]. The isolation of virus was also attempted in C6/36 cells from 28 acute phase samples (<5 days of onset of fever) following the standard virus adsorption technique [Reference Gould, Clegg and Mahy13].
The clinical history revealed that all the patients were suffering from fever (38·5–40°C), severe headache, body pain, fatigue, myalgia, and vomiting. Rash was observed in 30% of the patients. The serological analysis of the samples indicated 52·7% seropositivity with 12% IgM, 8·7% IgG and 32% with both IgM and IgG antibodies. A total of 15 (5·4%) serum samples were found positive for the presence of dengue-specific RNA through demonstration of 511-bp dengue complex-specific amplicons by RT–PCR. All these amplicons were further subjected to nested RT–PCR for serotyping which revealed nine as DENV-4 positive and six as DENV-3 positive. The details of PCR-positive samples are provided in Table 1. The three serial passages of 28 acute phase samples resulted in isolation of two DENV-4 and one DENV-3 isolates. The isolations were also confirmed by nested PCR and sequencing.
Table 1. Details of sex, age, antibody and serotype profile of dengue RT–PCR positive samples

The pair-wise nucleotide sequence comparison of complete envelope gene (1485 nucleotides) revealed that both the DENV-4 isolates from this outbreak were closely related (97% identity). These isolates revealed 95% identity with the prototype DENV-4 isolate (H-241), isolated from the Philippines in 1956. The phylogenetic analysis, based on the complete envelope gene, classified all the endemic/epidemic global DENV-4 viruses into three genotypes (Fig. 1). The sylvatic viruses were placed at the basal position of the tree [Reference Lanciotti, Gubler and Trent14]. Both Indian DENV-4 viruses (ND73 and ND110) from this outbreak were grouped into genotype I, which was represented by isolates from a large number of Asian countries including Thailand, Vietnam, Cambodia, Malaysia, the Philippines, China, and Sri Lanka. Within genotype I, the Indian DENV-4 viruses form a close branch with an isolate (GenBank accession no. AB111086), recovered in Japan. This virus was isolated from a viraemic Japanese traveller returning from India in 1996. A Sri Lankan isolate from 1978 (GenBank accession no. U18437), appears to be the progenitor of these Indian viruses. The close clustering of these three Indian viruses along with the Sri Lankan isolate of 1978 suggests the circulation of a unique clade of genotype I of DENV-4 in this geographical region. No other DENV-4 isolates from other geographical regions were grouped in this clade. Genotype II was represented by the majority of isolates from South America/Caribbean islands including Venezuela, Puerto Rico, Ecuador, Mexico, Brazil, Surinam, and Tahiti, which were found to be evolved from Indonesian DENV-4 viruses of late 1970. Surprisingly, genotype III was represented by only two DENV-4 isolates from Thailand, isolated in 1997. The phylogenetic tree drawn using NJ and MP also revealed a similar clustering pattern (data not shown).
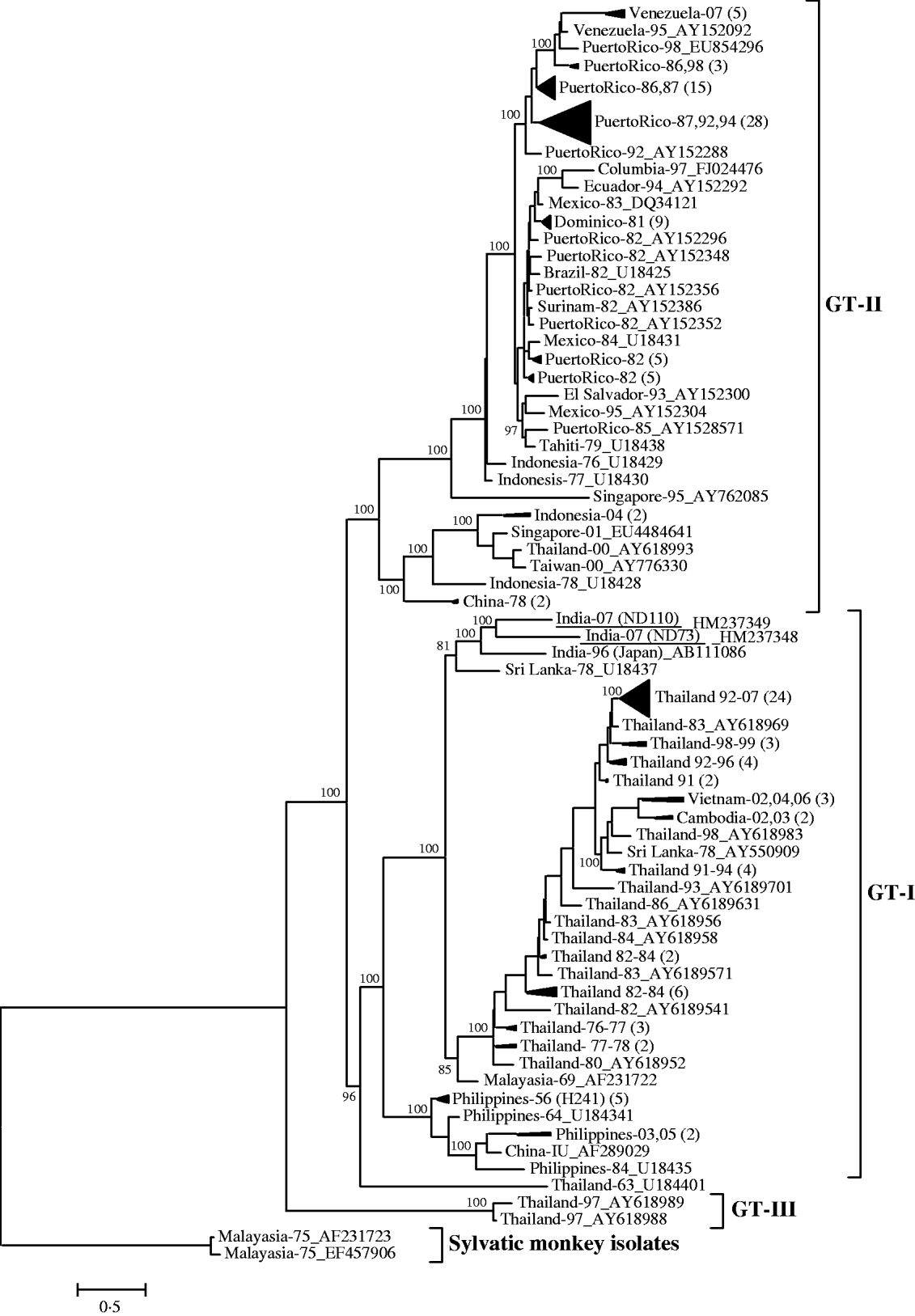
Fig. 1. Phylogenetic tree among DENV-4 viruses generated by Bayesian phylogenetic inference based on the nucleotide sequence of complete envelope gene (1485 nucleotides). Each strain is abbreviated with country of origin and last two digits of the year of isolation followed by the GenBank accession number. A group of phylogenetically closely related viruses from a specific country and specific year(s) are grouped into a cluster and designated by country of origin and last two digits of the year of isolation(s) followed by the number of isolates in parentheses. The DENV-4 strains sequenced in this study are underlined. Posterior probability values are indicated at the major branch points.
The nucleotide sequence analysis of the C-prM gene junction of six DENV-3-positive amplicons revealed close identity (>98%) with sequences of DENV-3 (genotype III) viruses circulating in India during 2003–2006. The phylogenetic analysis revealed a close branching pattern of these DENV-3 sequences with the existing Indian DENV-3 viruses (data not shown), indicating their continued circulation [Reference Dash9].
In the current study, the demonstration of dengue RNA in 5·4% of samples by RT–PCR and detection of IgM antibodies in 12% of samples confirmed the causative agent of this outbreak to be dengue virus. The isolation of DENV-4 and DENV-3 viruses from clinical samples further confirmed this aetiology.
Both the DENV-4 (ND73 and ND110) isolates in this study were found to be closely related (96·7%) to a DENV-4 isolated from a Japanese traveller who visited India in 1996 [Reference Ito15]. The Bayesian phylogenetic analysis also revealed their close branching pattern with 100% posterior probability support, indicating their common evolutionary origin. It can be presumed that DENV-4 viruses are circulating silently in India, although surprisingly DENV-4-associated major outbreaks have not been reported in last two decades. Many factors including inadequate human and entomological surveillance, low incidence or relatively lower vectorial competence of Indian Aedes mosquitoes for DENV-4 may be attributed to this paucity of transmission. The sudden emergence of DENV-4, displacing prevailing dengue serotypes is a point of concern, since the emergence of a new serotype in an area is often associated with severe and large dengue haemorrhagic fever (DHF) outbreaks [Reference Rico-Hesse16].
An epidemiological study in Thailand revealed that despite the low prevalence of DENV-4, it was responsible for 10% of DHF cases in children. Most of these DHF cases have been attributed to the secondary dengue infection with DENV-4 [Reference Nisalak17]. It has also been observed that although DENV-2 and -4 were introduced simultaneously into the Americas in 1981, the dispersal rate of DENV-4 was much higher. The population size of DENV-4 was estimated to have doubled in just 2 weeks compared to 32 weeks in DENV-2 viruses, indicating their higher epidemic potential [Reference Carrington18].
In summary, this is the first report regarding the emergence of DENV-4 (genotype I) in a dominant form in India. The genetic characterization of Indian DENV-4 viruses will serve as baseline information for future molecular epidemiological investigations. It will be interesting to track the dynamics of this unique clade of DENV-4 during future outbreaks.
ACKNOWLEDGEMENTS
The authors are grateful to Dr R. Vijayaraghavan, Director, Defence Research and Development Establishment, Gwalior, for providing the necessary facilities and financial grant for this study. The authors are also grateful to the Director, NIMS, Hyderabad for providing clinical samples.
DECLARATION OF INTEREST
None.




