1. Background
Neurodevelopmental and degenerative changes, addiction disorders and other psychiatric problems are both influenced by and mirrored in brain changes that occur throughout life. A major challenge is to determine which age-related changes are detrimental and which enhance cognitive and mental health. The potential economic benefits of an improved understanding are large, with total costs of brain disorders in Europe in 2010 estimated at €798 billion [Reference Gustavsson, Svensson, Jacobi, Allgulander, Alonso and Beghi1]. Throughout life, our genetic dispositions interact continuously with environmental, societal, occupational and lifestyle factors to influence brain structure and function. Such changes, from the earliest stages of life to oldest age, are mapped in detail in European longitudinal studies, utilizing Magnetic Resonance Imaging (MRI). MRI yields high-resolution images of variations in brain macrostructure, microstructure and function, which can be compared with measurable changes in cognitive function and mental health. However, since MRI is expensive and time-consuming, the number of participants included in such studies tends to be low. This makes it hard to disentangle the role of the many factors that can influence brain, cognition and mental health at different stages of life. While forming a precondition for a possible personalised medicine approach, such individual variations need first to be established. For instance, age-specific mechanisms necessitate a large number of participants at all stages of life, and sex-specific effects further halve the sample sizes, thus narrowing degrees of freedom available for analyses.
1.1. Overall aim and objectives
This EU Horizon 2020 project “Lifebrain” aims to maximise the exploitation of brain imaging cohorts by bringing together studies on how differences and changes in brain age relate to cognitive function and mental health. This will be done by integrating and standardizing data and results from 11 large predominantly longitudinal European samples from 7 countries [2–12] (Table 1).
Table 1 Central features of the studies that will feed into the Lifebrain database.

N: Number of unique individuals.
Obs: Total number of observations (one for every individual for each time point in longitudinal studies.
CM: Cognitive tests and mental health measures. MRI includes, T1, T2, DTI, RSI, resting-state fMRI, task-related fMRI. Proportion of participants with neurodegenerative disease varies.
* Number of unique subjects with MRI/total number of MRI sessions e.g. including follow-up; ** DNA: the genotyping varies some across studies. Many have GWAS, including all in NESDA, all in Base I and 300 in BASE II, ≈200 in LCBC, ≈2200 in Betula, and GWAS for 2000 will be performed as part of Lifebrain. Cam-CAN has collected saliva samples and DNA extraction and GWAS is a part of Lifebrain. All research participants described in Table 1.3 will be included in statistical analyses based on existing data. All participants that (a) can be reached, (b) are from samples where relevant ethical approval is given, (c) for whom relevant data do not already exist, and (d) are able to give full informed consent, will be asked for dried blood spot samples, buccal swab samples and online testing.
This will yield a database of fine-grained BCM health measures for more than 5.000 individual participants. Longitudinal brain imaging data are available for a major portion, as well as cognitive and mental health measures for broader cohorts, exceeding 27.000 examinations. The project is a collaborative initiative involving a small and medium-sized enterprise (SME), several of Europe’s major brain research centres, as well as stakeholders for efficient exploitation of results (Fig. 1).

Fig. 1 Lifebrain’s vision, mission, overall aim and main objectives, and their relationships.
Lifebrain includes four sub-objectives:
• Integration of data across existing major longitudinal European neuroimaging studies of age changes in brain, cognition and mental health, including genetic, epigenetic, lifestyle, and medical registry information for thousands of individuals, further enriched with health outcomes and biomarkers.
• Development and standardization of measures and methods across these major European studies of age changes in brain, cognition and mental health.
• Provision of novel information on brain, cognition and mental health maintenance, onset and course of diseases and health inequalities, to yield the evidence base for development of policy strategies for prevention and intervention, thereby addressing health inequalities.
• Communication and implementation of new knowledge, exploiting the integrative cohorts in age-specific prevention and treatment to optimise brain, cognition and mental health, improving clinical practice and health policy.
1.2. Vision
Personalized health care requires fundamental knowledge of risk factors and protective factors, as well as the pathways through which they work at different ages. Extrapolating from known effects of certain risks and interventions [13,14], a multifactorial and personalised approach could identify modifiable environmental factors that promote cognitive development in childhood and adolescence, foster maintenance of cognitive functions into late adulthood, delay onset of dementia, reduce need for care, and improve working ability through prevention and intervention programs. Cognitive and mental health disorders are a serious burden for individuals as well as societies [15,16]. Within 5–10 years, we hope knowledge established in Lifebrain will enable policy makers and health care systems to implement low-threshold strategies for individual prevention by modifiable life-style factors, as well as non-pharmacological interventions. In a European perspective, these could have enormous consequences for individual well-being, work abilities, and for the total costs related to increased health care needs and reduced working capabilities in older adults during the coming decades.
2. Methods
2.1. Concept
A new approach to model brain, cognitive and mental health is needed that differs in fundamental ways from previous approaches: it should be dimensional, focused on lifespan rather than specific phases of development or age, and based on systems-vulnerability and resilience, rather than simple cause-effect relationships. We argue, (1) factors that affect cognitive and mental health will often vary along a continuum across the population, (2) risks and benefits accumulate over time, and will not be coincident with the age at which their effects become apparent, and (3) the effects of these factors will vary across individuals as a function of their genotype. We aim to identify important causal factors, to improve our understanding of how these affect brain health at different ages and in different people, and to identify beneficial and cost-effective interventions. The project will proceed through distinct but tightly interacting phases (Fig. 2).

Fig. 2 Variable and analysis structure within Lifebrain.
We will further combine a large population-based approach with an in-depth neurocognitive approach in order to clarify mechanisms, and how these translate into specific cognitive functions. For instance, functional variation with age and sex across countries may be due to individuals in some regions of the world having experienced better conditions in childhood and adulthood, relating to nutrition, education, disease exposure and physical and social activity patterns [17,18]. However, the pathways and mechanisms by which these broad factors work, remain unknown. This knowledge is pivotal for development of efficient health policies and targeted prevention.
2.2. Identifying risk and protective factors
To identify risk factors for cognitive and mental disorders, we will use approaches previously developed to group participants according to their trajectories of change [19,20]. Factors such as higher education, physical activity, female sex, carrying the met-allele of the COMT gene, and not living alone were associated with maintenance of memory function; lower education, unemployment, being male, and carrying a APOE ε4 allele were risk factors for decline [Reference Josefsson, de Luna, Pudas, Nilsson and Nyberg20]. We have recently employed structural equation modelling (SEM) trees as a method to uncover heterogeneity in empirical data, together with predictors and potential interactions among them, explaining this heterogeneity [Reference Brandmaier, von Oertzen, McArdle and Lindenberger19]. We will use similar approaches to stratify larger numbers of participants according to important cognitive, psychiatric and brain health measures, and test the effects of a range of risk and protective factors, including genetics, epigenetics, socio-demographic, environmental, and lifestyle. This part of the project will allow identification of general risk factors for cognitive and mental health. Some of the relations will be correlational in nature; others can be modelled longitudinally to shed light on cause and effect. Pathways for the observed relations, and how they are moderated by a number of other factors, will be specifically targeted (see below).
We will also assess long term effects of candidate variables from early life. The earliest of these are markers of pre- and neonatal health. Abnormal cognitive and behavioural outcomes, such as in ADHD and schizophrenia, have been linked to early development, including factors related to foetal growth and adversity [21,22]. Low birth weight, as a marker of adverse intrauterine circumstances, has been associated with a range of diseases and reduced function in daily life [23,24]. Some factors are associated with cognitive decline and increased Alzheimer's disease (AD) risk, e.g. coronary heart disease, hypertension, and type 2 diabetes [Reference Syddall, Sayer, Simmonds, Osmond, Cox and Dennison25]. Anatomical studies show that low birth weight, premature birth, and prenatal substance exposure can affect cortical and specifically fronto-striatal development, impacting attention and executive function [26–28]. These findings underscore the importance of neonatal characteristics for brain development and individual functional differences along a continuum from normality to pathology [Reference Walhovd, Tamnes and Fjell26].
Adding to this picture, a number of common variants in risk genes for psychiatric disorders were recently found predictive of brain structure at birth [Reference Knickmeyer, Wang, Zhu, Geng, Woolson and Hamer29]. For instance, neonates carrying APOE ε4, the major genetic risk factor for AD, were reported to have reduced volumes of temporal cortex, similar to that reported in older adults [Reference Knickmeyer, Wang, Zhu, Geng, Woolson and Hamer29]. For variants of the fat mass and obesity-associated gene (FTO), which has been related to reduced brain volumes in healthy ageing and risk of AD [30,31] and depression [Reference Milaneschi, Lamers, Mbarek, Hottenga, Boomsma and Penninx32], smaller brain volumes were recently shown also in adolescents [Reference Melka, Gillis, Bernard, Abrahamowicz, Chakravarty and Leonard33]. Such findings should increase our attention to researching and optimizing early influences to provide resources and knowledge on health and disease determinants, onset and course of diseases and public health. Efforts to postpone decline or disease may be futile if they are only targeted at old age. By including the full lifespan, we will investigate how early life variables, including genetic and epigenetic variants, exert their effects.
In addition to the very early predictors related to pre- and perinatal conditions, we will also focus on indicators in childhood and young adulthood. For instance, intelligence scores at age 11 are remarkably good predictors for cognitive performance at age 90 years [Reference Deary, Pattie and Starr34], and neuroticism and introversion predict development of depression later in life [Reference Hakulinen, Elovainio, Pulkki-Raback, Virtanen, Kivimaki and Jokela35]. Furthermore, cardiovascular fitness and cognitive performance in early adulthood increased the risk of early-onset dementia and mild cognitive impairment (MCI) more than four decades later [Reference Nyberg, Aberg, Schioler, Nilsson, Wallin and Toren36]. Current levels of self-reported physical activity have been related to less cortical atrophy in the prefrontal cortex across a 3.5-year period in the adult lifespan [Reference Walhovd, Storsve, Westlye, Drevon and Fjell37]. This may suggest that older adults should exercise more to prevent cortical atrophy. Alternatively, the more physically active older adults may also have been the more physically active and fit younger adults. Conversely, certain brain characteristics or changes may affect people’s physical activity [Reference Sabia, Dugravot, Dartigues, Abell, Elbaz and Kivimaki38].
It is pivotal to examine to what extent the late-life association can be explained by early physical fitness, current physical activity, and their relationship, or if both exert unique influences. People with body mass index (BMI) >25, showed a relationship between BMI and brain atrophy [Reference Walhovd, Storsve, Westlye, Drevon and Fjell37]. However, for older adults, higher BMI may not confer the same risk as for younger or middle-aged adults, and may even be protective, conferring a smaller risk of cardiovascular complications [Reference Batsis, Singh and Lopez-Jimenez39]. Similarly, certain genetic variations, such as the APOE ε4 allele, reduce brain plasticity and increase brain vulnerability [Reference Sundstrom, Marklund, Nilsson, Cruts, Adolfsson and Van Broeckhoven40]. Thus, we might expect less stability of cognitive function [Reference Josefsson, de Luna, Pudas, Nilsson and Nyberg20] and a higher vulnerability to decline in the face of other risk factors for those with the risk allele. Equally, brain-derived neurotrophic factor (BNDF) is linked to neuronal growth and differentiation, and thus contributes to memory and learning, leading to differential cognitive trajectories [Reference Ghisletta, Backman, Bertram, Brandmaier, Gerstorf and Liu41] and to mental health differences [Reference Hosang, Shiles, Tansey, McGuffin and Uher42]. These findings are in line with the broader “resource modulation” hypothesis [Reference Lindenberger, Nagel, Chicherio, Li, Heekeren and Backman43], according to which the effects of common genetic variation on cognitive performance increase from early to late adulthood, reflecting the non-linear association between brain resources and performance (for review, see [Reference Papenberg, Lindenberger and Backman44]).
In Lifebrain, using integrated cohorts representing diverse European social models over a large age range, and with the additional use of US databases, we will test national differences in risk and protective factors in BCM health.
A critical aspect of Lifebrain is the unique longitudinal neuroimaging design in some of its cohorts. Data from the first six time-points of the HUBU-project, for example, show that the developmental trajectories in hippocampal volume are not well predicted from only cross-sectional observations using a generalized additive model (left panel of Fig. 3), compared to when estimated from longitudinal information (with a generalized additive MIXED model, right panel), which takes actual change into account. The same phenomenon is frequently observed in ageing, where unavoidable recruitment bias may lead to under-estimation of true age effects.
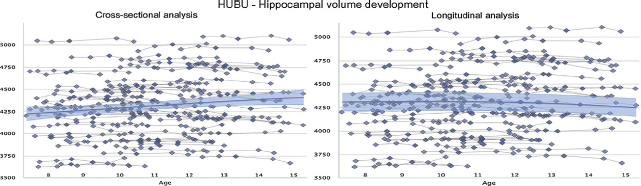
Fig. 3 Developmental trajectories for hippocampal volume are very different when estimated from cross-sectional observations only by a generalized additive model (left panel of figure below) compared with a generalized additive MIXED model (right panel) taking actual change into account [10,11]. Hippocampal volume [mm3] was measured from T1-weighted MRI images, segmented by FreeSurfer from the left hemisphere. The fit curves were based on 510 scans from 94 participants (mean age 11.2 years, 7.5–15.4 years).
In summary, we will analyse the entire lifespan, including early life characteristics, and individual factors, as well as the level of the individual and broader society, and identify risk and protective factors for brain, cognition and mental health. We will achieve this by combining population-based, cross-sectional and longitudinal cohorts with general indicators of health, together with in-depth detailed and targeted cognitive analyses across European and US studies.
2.3. Identifying pathways and moderators
Our longitudinal design permits identification of pathways from risk and protective factors to cognitive and mental health problems and their moderators that affect the strength or direction of relationships. We will examine how broader risk factors work at a biological and behavioural level (pathways), and evaluate interactions between identified risk factors, intermediate variables as part of pathways, and along with other important variables, e.g. sex [45,46]. Sex has been associated with differences in the risk for developing several disorders [45–48]. Observed sex differences in behaviour, cognition, emotions, and measures of brain structure and function, and their development [45,49,50] are often subtle and inconsistent, as many studies investigate limited age ranges or lack longitudinal observations [Reference Mendrek51]. Moreover, the effect of sex may differ across the lifespan [20,37,52,53]. Improved living conditions in Europe and less gender-restricted education are associated with increased sex differences in some cognitive functions, such as episodic memory, favouring women, and also decreases of others, such as numeracy [Reference Weber, Skirbekk, Freund and Herlitz18]. It is interesting whether pathways at the level of brain and other biomarkers can be identified for sex as a moderator of cognitive and mental health over time, and across physical and social environments, including countries. Variables of interest include fatty acids, BMI, vitamins, cholesterol, blood pressure, genetic and epigenetic factors, and MRI measures of the brain. Outcome measures in Lifebrain will include many objective health variables, and refined cognitive and brain imaging assessments, mental health measures, as well as social and mental factors. This will be used to address the pathways and mechanisms for identified risk and protective factors. Essential fatty acids are important for development of cognitive function in normal pregnancies [Reference Helland, Smith, Saarem, Saugstad and Drevon54], as well as for severely premature infants [Reference Henriksen, Haugholt, Lindgren, Aurvag, Ronnestad and Gronn55]. BMI and certain blood markers are, for example, related to less (e.g. DHA, vitamin D) or more (e.g. cholesterol) brain atrophy [Reference Walhovd, Storsve, Westlye, Drevon and Fjell37]. Vitamin D has received much attention lately, as the link between vitamin D deficiency and risk of mental and neurodegenerative diseases has been established [32,56], and insufficiency or deficiency of vitamin D is common in older clinical samples. Little is known about the direction of the relationship, and whether similar relationships can be found among groups of non-demented elderly [Reference de Koning, van Schoor, Penninx, Elders, Heijboer and Smit57]. Higher levels of vitamin D appear to be related to less degeneration of major white matter tracts in the brain, even in cognitively healthy elderly participants. Vitamin D may stimulate production of neurotrophic, antioxidative and anti-inflammatory factors, reduce risk for cardiovascular and cerebrovascular disease and even influence amyloid phagocytosis and clearance [Reference Etgen, Sander, Bickel, Sander and Forstl56]. If these relationships between brain change and nutrients like vitamin D can be replicated across larger and international samples, such mechanisms will provide a foundation for targeted interventions and randomized controlled trials also in normal ageing.
We do not know how potentially modifiable behaviours, e.g. eating habits and physical activity, affect blood nutrient and vitamin levels. Also unknown are effects of modifying medical factors such as BMI and hypertension, how genetic factors interact, and the impact of early-life cognitive function. There is reason to expect that established genetic variants, e.g. FTO [Reference de Luis, Aller, Conde, Izaola, Gonzalez Sagrado and Castrodeza Sanz58], may influence the relation between food intake and blood nutrient and cardiovascular markers, and also the relation of such markers with brain atrophy. Furthermore, APOE ε4 may reduce brain plasticity, and increase vulnerability to negative impact on cognitive function [Reference Sundstrom, Marklund, Nilsson, Cruts, Adolfsson and Van Broeckhoven40], as well as mental health [Reference Skoog, Waern, Duberstein, Blennow, Zetterberg and Borjesson-Hanson59]. APOE ε4 carriers also have more pronounced sleep disturbances [Reference Hita-Yanez, Atienza, Gil-Neciga and Cantero60] and poor sleep habits have been related to brain atrophy [Reference Sexton, Storsve, Walhovd, Johansen-Berg and Fjell61], poor white matter integrity [Reference Sexton, Zsoldos, Filippini, Griffanti, Winkler and Mahmood62] and amyloid beta deposition [Reference Branger, Arenaza-Urquijo, Tomadesso, Mezenge, Andre and de Flores63] in aged individuals. Positive lifestyle factors such as physical activity [Reference Brown, Peiffer, Taddei, Lui, Laws and Gupta64], healthy diet [Reference Gardener, Rainey-Smith, Barnes, Sohrabi, Weinborn and Lim65] and high education levels [Reference Wang, Gustafson, Kivipelto, Pedersen, Skoog and Windblad66] may be beneficial for APOE ε4 carriers, by lowering amyloid deposition [Reference Vemuri, Lesnick, Przybelski, Knopman, Machulda and Lowe67] or promoting ‘maintenance' of brain metabolism [Reference Arenaza-Urquijo, Gonneaud, Fouquet, Perrotin, Mezenge and Landeau68]. Such observations are examples of considering distinct lifestyles in the context of a single or combination of genetic variant(s), in order to develop tailored strategies for intervention in subgroups of older adults. We will measure blood-based nutritional factors, food intake, genetics, and epigenetics, and relate them with measures of brain and cognitive function. Our combination of samples is vital to obtain sufficient statistical power.
The rich information in our large longitudinal databases will allow us to examine individual differences in the onset and magnitude of cognitive change, and identify genetic and lifestyle factors that predict preserved or declining cognition. In the Betula study [Reference Josefsson, de Luna, Pudas, Nilsson and Nyberg20], there were three distinct ageing trajectories for episodic memory (Fig. 4).

Fig. 4 By tracking memory performance over 15 years, factoring in attrition, we were able to show that 18% of 1558 participants upheld good memory function in ageing, while 13% declined [Reference Josefsson, de Luna, Pudas, Nilsson and Nyberg20]. The memory score was a composite based on 5 episodic-memory tasks (max score = 76).
Good episodic memory in old age was also associated with preserved hippocampal function [Reference Pudas, Persson, Josefsson, de Luna, Nilsson and Nyberg69]. Parkinson's disease patients with MCI have reduced fronto-striatal performance of working memory [Reference Ekman, Eriksson, Forsgren, Mo, Riklund and Nyberg70]. It is likely that those older participants displaying fronto-striatal/working memory impairment will be distinct from those showing hippocampus/episodic memory impairment, and that distinct genetic and lifestyle predictors are relevant for each phenotype. The same may apply to the separation of verbal and visuo-spatial memory [Reference Suri, Topiwala, Filippini, Zsoldos, Mahmood and Sexton71]. Identification of specific cognitive phenotypes may suggest that different kinds of interventions will be effective. In addition to the broad set of existing genetic, epigenetic, health, demographic and lifestyle variables available in the Lifebrain database, we will conduct additional analyses to enrich the database with, e.g. telomere length [Reference Wikgren, Karlsson, Nilbrink, Nordfjall, Hultdin and Sleegers72] and longitudinal brain imaging [Reference Nyberg, Salami, Andersson, Eriksson, Kalpouzos and Kauppi73].
In summary, we will identify variables and pathways with impact on neurocognitive function and mental health throughout life to be used in targeted intervention and prevention.
2.4. Standardization and harmonisation
Standardization of measures across sites post hoc, as well as through enrichment of existing cohorts, is enacted in two separate work packages (WP). Partners have experience with multi-site neuroimaging, cognition and mental health studies. We will utilize procedures developed and described by the BioSHaRE project for data harmonisation, integration and federated data analyses [Reference Doiron, Burton, Marcon, Gaye, Wolffenbuttel and Perola74]. Measures can also be harmonized across latent variables: Since existing data have not been collected with identical scales and measures (e.g. different versions of neuropsychological tests and different MRI scanners), site-specific differences are expected for some measures. To allow measurements to be made more resistant to measurement error, and to abstract from individual scales to underlying constructs, our analyses will therefore be based also to some degree on latent factor models. We will employ standard psychometric approaches to determine construct validity across sites. We will rely on the standardized items of the new data collection phase. If standardized items are collected as a partial reassessment of site-specific items from the original data collection, this allows for the evaluation of convergent and divergent construct validity. We will test the convergence of latent factors from site-specific and standardized instruments to determine statistical overlap of hypothetical constructs across sites. This is also an effective means of dealing with missing data. Such an approach will allow merging of data across sites, and will probably be beneficial in multi-site studies outside the collaboration.
2.5. Linkage with registry data
For some of the Lifebrain data, linkage to registry data may be possible. Indeed, some have already been linked by individual consent – e.g. the MRI cohorts from the Norwegian Mother and Child Study (MoBa) are linked, and Norwegian adult cohorts also have consented to link their MRI, cognitive, mental health and genetic data with the Medical Birth registry, as well as Army conscription data. Further consent for individualised linkage with population registries will be pursued especially for some Scandinavian samples.
2.6. Critical measures
2.6.1. Magnetic resonance imaging (MRI)
MRI at all sites includes T1-weighted scans for morphometric analyses. In addition, many cohorts have diffusion-weighted scans for structural connectivity and resting-state BOLD-weighted scans for functional connectivity analyses. Diffusion tensor imaging (DTI) in particular is sensitive to microstructural brain tissue properties, and is a promising biomarker related to development, ageing, disease, and cognition [Reference Amlien and Fjell75].
2.6.2. Neuropsychology
Extensive cognitive tests are available for all cohorts, with validated and reliable measures. We focus especially on general cognitive abilities and episodic memory, in addition to cognitive rating scales and measures of daily life function. With some variation across cohorts, cognitive measures will also include working memory, executive function and processing speed.
2.6.3. Genetics and epigenetics
Saliva or blood for extraction of DNA have been collected for all but two of the samples included in Lifebrain. In addition, we will examine about 2000 further individuals.
2.6.4. Dried blood spots (DBS)
DBS will be collected at most sites and analysed by Vitas (WP2 and WP3). Priority will be given to nutrients, such as essential fatty acids, cholesterol, and vitamins. Vitas will develop and validate new biomarkers on DBS tailored for Lifebrain. Candidates for new biomarkers are specific proteins, and from the Vitas comprehensive UPLC-HR-TOF-MS lipidomics platform, a panel of lipid markers with potential to predict preclinical transition to clinical stages of Alzheimer's disease.
2.6.5. Lifestyle and health measures
For most participants, information about lifestyle and general health is available, such as blood pressure, recordings of medication and other indices of cardiovascular disease and BMI, and will be enriched in a harmonized way through online data collection. Data on sleeping problems exist for most subjects and will be enriched through online data collection.
2.6.6. Mental health
Standardized measures of symptoms of depression and anxiety exist for most participants and will be enriched in a harmonized way through online data collection.
2.7. Investigations
Our approach calls for sophisticated modelling, and experts in longitudinal statistical modelling, bioinformatics, and genetics are included in the core group, as well as renowned neuroimaging experts. A specific task is dedicated to build an integrated multi-modal processing stream for Lifebrain, based on combining existing tools (FreeSurfer and FSL) with custom made procedures and new developments.
2.7.1. Neuroimaging preprocessing and statistics
FreeSurfer (FS) will be used for quantification of cortical thickness, volume and local arealization continuously across the brain surface, and volumes of a range of subcortical structures, including hippocampus. FS segments images at sub-voxel resolution and detects fine-graded effects, validated by manual segmentations and histology. All cortical metrics will be sampled in the same surface-based template, yielding unique possibilities for multi-modal integration by combining a range of different metrics while ensuring spatial coherence.
For DTI we will combine two approaches. We will use tract-based spatial statistics (TBSS) developed in the FMRIB Centre at Oxford for whole-WM comparisons. This allows voxel-based comparisons that are robust to anatomical differences and partial volume effects. For quantification of changes in major WM tracts, we use a newly developed automated and robust probabilistic reconstruction scheme – Tracula (TRActs Constrained by UnderLying Anatomy) [Reference Yendiki, Reuter, Wilkens, Rosas and Fischl76]. It is especially suited for longitudinal analyses, with high sensitivity and no bias in change estimates.
2.7.2. Cross-site standardization of neuroimaging data
The MRIs come from different scanners, which will affect the absolute measurement values. The applicants have extensive experience with large multi-centre neuroimaging, including ENIGMA [Reference Hibar, Stein, Renteria, Arias-Vasquez, Desrivieres and Jahanshad77], ADNI, e.g. [78,79], PING [27,28], and the Oslo Multi-Sample Aging Study [Reference Fjell, Westlye, Amlien, Espeseth, Reinvang and Raz80]. The segmentation procedures used in this project are not biased by scanner platform and seem not to affect the strength of relationships to neuropsychological scores [Reference Han, Jovicich, Salat, van der Kouwe, Quinn and Czanner81]. Atlas-based normalization to increase robustness and accuracy of the segmentations across scanner platforms [Reference Han and Fischl82], and normalizing analyses for differences in grey matter – white matter contrast will increase sensitivity in multi-site studies [Reference Douaud, Refsum, de Jager, Jacoby, Nichols and Smith83]. Also, the size of each subsample will be large enough to allow statistical adjustment by scanner. Internal validation in the Oslo Multi-Sample Aging Study, showed a minor decrease in sensitivity by pooling together data from six different samples and scanners was accompanied by a large increase in power. In summary, we are well suited to tackle the challenges inherent in any multi-site study, and benefit from the manifold increases in sample size. As a feasibility check, we compared the age-trajectories of hippocampal volume from the Cam-CAN sample (n = 651) and an LCBC subsample (n = 1100) and observed highly comparable trajectories.
2.7.3. Genetic and epigenetic analyses
Data generation for genome- and epigenome-wide association study (GWAS and EWAS) analyses will be coordinated by University of Lübeck, using state-of-the-art high-throughput genome technologies for microarray-based profiling of DNA sequence (for GWAS) and methylation states (for EWAS). Several datasets have genome-wide SNP data available, and in addition we will select 2000 individuals from at least four different datasets for de novo collection of buccal swabs, allowing us to determine both genome-wide SNP genotype and DNA methylation profiles using the “Global Screening Array” and “Infinium Methylation EPIC” array, respectively (both Illumina Inc.). Buccal swabs will be collected by participants at home using the Catch-All Sample Collection Swabs (Epibio, Inc.) following standardized collection protocols. Genome-wide data will serve several purposes in Lifebrain: First, the existing genotype and newly generated DNA methylation data can be used in the context of GWAS and EWAS analyses, to identify novel (and confirm previously reported) genetic/epigenetic determinants – and their interaction – of the cognitive and imaging traits of interest. Second, the genome-wide genotype data allow to precisely assess and correct for subtle differences in population substructure within and across cohorts allowing to optimise data analysis procedures and inferences of non-genetic variables.
2.8. Data integration and statistical considerations
2.8.1. Statistical modelling of change
The complex longitudinal nature of the data poses significant statistical challenges in order to fully exploit the potential of combined data. To this end, Lifebrain has dedicated a task to development of new statistical tools. The consortium will address statistical challenges in three steps: (1) comparative analysis of data sets and research designs; (2) development and application of statistical tools; (3) tool refinement and model selection: For Step 1, the different research designs represented in Lifebrain will be compared to obtain an overview of their relative strengths, including differences in statistical power to detect effects of interest. Comparative analysis will inform data analysis strategies and future expansion of data sets. We will introduce a statistical tool that permits researchers to compute effect sizes that allow for unequal and person-specific measurement intervals, non-linear change, and selective attrition. For Step 2, we will promote the application of three interrelated sets of statistical tools: multivariate and dynamic variants of longitudinal structural equation modelling (SEM); classification and regression trees (CART); generalized additive mixed modelling (GAMM). Multivariate dynamic SEM is well suited to identify lead-lag relations among constructs representing brain functions and structures, cognitive performance, and health outcomes. CART and related data mining techniques help to uncover classes of individuals with similar profiles and identify relevant predictors of class membership even with very large numbers of interacting predictors. GAMM offers powerful tests of nonlinear effects on univariate criterion variables, including interactions with other covariates. We will introduce two new tools that combine the benefits of SEM and CART: (i) SEM trees [Reference Brandmaier, von Oertzen, McArdle and Lindenberger19] for discovering formerly undetected subgroups that differ in SEM parameters; (ii) SEM forests [Reference Brandmaier, Prindle, McArdle and Lindenberger84] for identifying variables that excel in predicting individual differences in such parameters across many predictors. We will also combine the benefits of each of the three approaches by proposing simultaneous estimation techniques for generalized multi-level models and evaluate the conditions under which they surpass existing stepwise procedures. In Step 3, we will refine the tools made available in Steps 1 and 2 based on feedback from research throughout the project. Furthermore, we will tackle the important problem of identifying and selecting those models that best summarize data across sites. We will adapt split-sample schemes and information-theoretic measures to control for multiple testing and avoid overoptimistic inferences due to double-dipping. The goal is to identify models that optimally represent the consolidated findings of the consortium, and hence contribute to theory development and generalizability. Members of the group have developed statistical tools for estimating the statistical power to detect individual differences in change in the context of longitudinal studies analysed with structural equation or multi-level models, such as latent growth curve models [85–89]. Use of these tools will permit group members to check whether the power to detect moderators of change with a given data set is adequate, and inform future design decisions, such as the spacing of measurements, or recruitment of new cohorts. In general, we expect the large databases of high-quality, well-validated and in-depth measures included in Lifebrain to yield excellent statistical power.
2.8.2. BIG data – storage, transfer and processing system
Sharing of brain imaging data between large cohort studies across Europe for integrative and comparative analyses is a core of Lifebrain research. Lifebrain will establish an international brain image sharing and analysis platform. For this, we will build on expertise gained through existing collaborative data platforms among the participants (i.e., the Dementias Platform UK Imaging Informatics, DPUK-II; UOXF; UK Biobank, MRC; and the Max Planck Brain Imaging Library, OpenBILD, MPIB).
Integrated analyses of the data are envisioned in two forms: (a) fully integrated pre-processing and analysis of raw imaging data (mega-analysis); (b) statistical integration of local pre-processing and analysis (meta-analysis). A fully integrated analysis of raw data would probably achieve the best possible integration. However, formal and technical constraints might prevent sharing raw data from some sites, so the integration of locally processed data (e.g., shared in tabulated form) will be considered.
At least for the moment, the ethical permission of the different participating centres is restricted to sharing within the Lifebrain consortium. As these permissions require to be specific and are slightly different for different countries, some data sets will be available to external applicants as a condition of funding (e.g. UK MRC), while for others a post hoc ethical approval for sharing is required and will be tied to specific conditions.
2.9. Uptake of research outputs
Lifebrain will develop mechanisms and tools to engage various stakeholders and bring their views and priorities to the project. Scientific exchange will take place with relevant policy makers, national decision makers, healthcare providers, patient organizations, cohort participants and researchers to support the uptake of project outputs. These outputs will be translated into specific guidelines and recommendations that will be actively disseminated according to the consortium’s dissemination, exploitation and communication plan
3. Conclusions
By integrating and standardizing major longitudinal studies of brain, cognition and mental health, Lifebrain aims to maximise the potential of European brain imaging cohorts. This will facilitate identification of the key determinants of cognitive and mental health across all ages, from birth to old age. Our lifespan focus fits with the novel life-course model of risk recently published by the Lancet Commission on Dementia prevention, intervention and care [Reference Livingston, Sommerlad, Orgeta, Costafreda, Huntley and Ames90]. While we do not specifically target old age and dementia, identification of the risk factors and protective factors at all stages of life will be critical to enable future prevention of cognitive and mental disorders.
Acknowledgement
This research is funded by the EU Horizon 2020 Grant: ‘Healthy minds 0–100 years: Optimising the use of European brain imaging cohorts (“Lifebrain”)’. Grant agreement number: 732592. Call: Societal challenges: Health, demographic change and well-being.








Comments
No Comments have been published for this article.