1. INTRODUCTION
Biologically inspired design (BID) is the discipline where inspiration is taken from the natural world to solve technical problems. BID is receiving increasingly more attention from research and industry because of the two main advantages the field is often associated with: sustainability and proven performance (Benyus, Reference Benyus1997; Bar-Cohen, Reference Bar-Cohen2011). Furthermore, drawing inspiration from a largely unused biological knowledge domain entails a higher probability of identifying leapfrog innovations. Other noteworthy advantages of biomimetic products are their enhanced marketability and financial savings through efficient use of energy and other resources.
These high expectations of biomimetic products are currently not met with adequate methods and algorithms that enable designers to systematically identify candidate biological strategies for biomimetic design. Most existing biomimetic ideas currently originate from spontaneous inspiration. For example, George de Mestral, the inventor of Velcro, serendipitously observed the ability of the cocklebur to attach to the fur of his dog. This inspired him to study the phenomenon in detail and to develop the well-known innovation. Another way to integrate bioinspiration into the innovation process is the employment of a multidisciplinary design team. This approach is expensive and provides no guarantee for success because biologists are typically specialized and hence biased to their specific field of expertise.
As the objective for the research underlying this paper, a scalable BID ideation system is envisaged that leverages the world's knowledge about nature and identifies those biological strategies that are interesting for a specific design problem. Currently about 1.7 million species are named, but the total number is expected to be 5–30 million (Purves et al., Reference Purves, Sadava, Orians and Heller2001). Although today only a fraction of these 1.7 million identified organisms is studied in detail, there exist many sources, such as books, journals, and online resources, where biological knowledge is documented. Considering the large work that lays ahead for biologists to completely describe and comprehend all of nature's phenomena, these sources are expected to keep on growing. The proposed approach is based on AskNature (http://www.asknature.org), a free to use, online bioinspiration tool built on a three-level, hierarchical classification, called the Biomimicry Taxonomy, to structure its database. It is the manual positioning of individual biological strategies into this information structure that currently limits AskNature to scalably integrate large numbers of biological strategies. This paper presents an automated classification approach that eliminates this time-consuming task and test results indicating the feasibility of realizing a scalable bioinspiration system.
2. NOMENCLATURE
Biomimicry Taxonomy: A three-level, hierarchical classification mechanism developed to organize AskNature's knowledge base
Class: a function-level classification category of the Biomimicry Taxonomy
Class support: the number of reference documents in AskNature's database for a specific Biomimicry Taxonomy class
Class weight: calculated as the maximum support divided by the specific class's support
Corpus: a collection of biological strategies in natural-language format
Part of speech: a linguistic category of words, for example, verbs, adjectives, nouns
Reference document: a biological strategy from AskNature's database used to train the proposed algorithm
Sample document: any biological strategy described in natural-language format
Sample score for a class: the number of k-nearest neighbor (k-NN) reference documents with reference classification to a specific class
Weighted score: the product of the sample score and the class weight
3. RELATED RESEARCH
This section provides an overview of related research with the emphasis on the integration of large numbers of biological strategies in the systematic BID process. The authors provide more detailed descriptions in Vandevenne et al. (Reference Vandevenne, Verhaegen, Dewulf and Duflou2011), where each contribution is positioned in the following four BID process steps: problem formulation, solution search, filter and analysis of alternatives, and knowledge transfer.
A manual, iterative bioinspiration search (Lenau et al., Reference Lenau, Dentel, Ingvarsdóttir and Guðlaugsson2010), starts from a functional keyword search and, from the obtained results, extracts new biological keywords for future searches. In this way, biological search words, initially not known to be relevant to the problem, are identified. A contribution aimed at automating the identification of biologically relevant search words is proposed by Chiu and Shu (Reference Chiu and Shu2007) and Shu (Reference Shu2010). The method aims at bridging the terminology gap between the engineering and biological domain by means of a systematic, semiautomatic search method that requires the design problem to be expressed in functional keywords and then generates biological meaningful bridge verbs and text passages containing them.
There are three model-based approaches that require the manual instantiation of detailed models for each biological phenomenon to be integrated in a structured knowledge base. Such a methodology has currently been reported for structure–behavior–function (SBF) models (Vattam et al., Reference Vattam, Wiltgen, Helms, Goel and Yen2010; Goel et al., Reference Goel, Vattam, Wiltgen and Helms2012), for functional basis models (Nagel et al., Reference Nagel, Nagel, Stone and McAdams2010; Nagel & Stone, Reference Nagel and Stone2012), and for state change, action, part, phenomenon, input, organ, and effect (SAPPhIRE) models of causality (Chakrabarti et al., Reference Chakrabarti, Sarkar, Leelavathamma and Nataraju2005; Sartori et al., Reference Sartori, Pal and Chakrabarti2010). These three model-based approaches have been recently extended in the following ways. In order to scale the SBF approach, Biologue, a social citation cataloguing system is developed (Vattam & Goel, Reference Vattam and Goel2011) to involve more people in the process of manual creation of SBF models. The functional basis approach is extended with an Engineering to Biology Thesaurus (Cheong et al., Reference Cheong, Chiu, Shu, Stone and McAdams2011; Nagel & Stone, Reference Nagel and Stone2012), a lookup table that translates the functional basis terms into biological corresponding terms. Finally, the SAPPhIRE approach is extended by an ontology aimed at providing extra stimuli during bioinspired ideation. The ontology consists of manually derived, biological and engineering term clusters for each of the SAPPhIRE model constructs (Srinivasan et al., Reference Srinivasan, Chakrabarti and Lindemann2012).
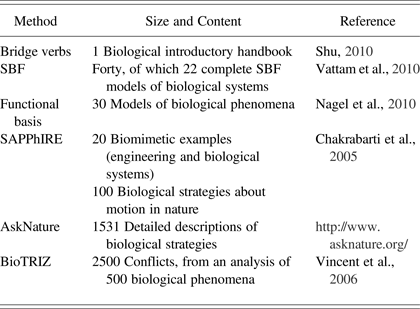
The following two contributions require positioning each biological strategy into a classification scheme. First, AskNature places biological strategies in a functional, hierarchical taxonomy called the Biomimicry Taxonomy. The designer looking for bioinspiration needs to formulate his or her design problem in this taxonomy. Second, BioTRIZ (Vincent et al., Reference Vincent, Bogatyreva, Bogatyrev, Bowyer and Pahl2006) aims at integrating biological knowledge in the TRIZ methodology (Altshuller, Reference Altshuller1984) by positioning biological strategies in the BioTRIZ contradiction matrix. To identify bioinspiration, the problem needs to be formulated into a classical TRIZ contradiction, which is then reformulated into a BioTRIZ contradiction. This BioTRIZ contradiction then leads the designer to inventive principles learned from the manual analysis of 2500 contradictions in 500 biological phenomena. In order to illustrate the resources integrated into the above systems, for each contribution, the number of reported biological sources is given in Table 1.
Table 1. Overview of existing database sizes and content

Note: SBF, Structure–behavior–function; SAPPhIRE, state change, action, part, phenomenon, input, organ, and effect.
All of the above methodologies struggle in one way or another with scalably leveraging large numbers of biological resources in natural-language format. Both the iterative bioinspiration search and the bridge verbs-based search entail extensive manual result filtering. All model-based approaches are difficult to scale because they require a detailed analysis of both the engineering and the biological systems to express them on a common abstraction level. Their thesaurus or ontology extensions raise questions about the completeness of the relatively short biological word lists, which, in turn, makes it difficult to estimate how much of the biological inspiration in natural-language texts can be retrieved when scaling these methodologies to large biological corpora; and validation is typically reported on a small number of handpicked cases. Crowd sourcing, recently reported for the SBF model-based approach (Vattam & Goel, Reference Vattam and Goel2011), in theory, could tackle the scalability of any BID ideation system. However, the successful creation of a large number of SBF models by an online community has not yet been reported. Third, the two classification-based approaches require the classification of each new biological strategy. This manual task translates to the positioning of the strategies into the Biomimicry Taxonomy, for the AskNature approach, or to the manual identification of the relevant contradiction, for the BioTRIZ approach. These are again tasks that demand interactive work proportional to the size of the biological databases. Like the SBF model-based approach, AskNature relies on an online community to aid in database expansion. The authors have observed the AskNature database to grow with about 100 strategies a year over the last 3 years. Although this number seems small compared to the total number of biological strategies that are being documented by humans, AskNature's database is the only initiative known to the authors that shows any significant and consistent database growth. The research presented in this paper details an approach to automatically populate the Biomimicry Taxonomy and hence scale AskNature's bioinspiration approach.
4. AskNature AND THE BIOMIMICRY TAXONOMY
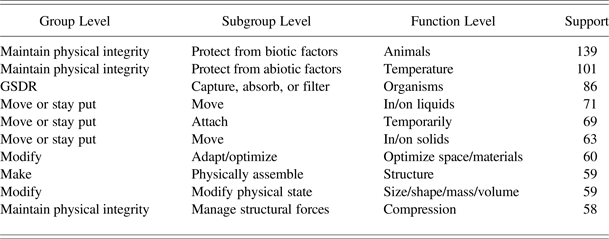
AskNature, developed by the Biomimicry 3.8 Institute (http://www.biomimicryinstitute.org), is an online tool that provides support for designers during concept generation in the early stages of the BID process. To use AskNature, a designer is required to formulate his problem into AskNature's Biomimicry Taxonomy, a functional, three-level hierarchical classification mechanism developed to organize his knowledge base. A small excerpt from the Biomimicry Taxonomy is presented in Table 2. The first level is the group level, which consists of eight categories, for example, “move or stay put.” Each group-level category is divided into subcategories named subgroups. The two subgroups for the “move or stay put” group are, for example, “attach” and “move.” Subgroups are further divided into functions. For example, “attach temporarily” and “attach permanently” are two function categories for the subgroup “attach.” In this way, the biological strategy of the octopus using suckers to attach itself is, for example, classified across the three levels as: “move or stay put” / “attach” / “attach temporarily.” Once the person looking for inspiration from nature has formulated his or her problem into the Biomimicry Taxonomy, AskNature returns a list of biological strategies that are previously manually classified into the chosen Biomimicry Taxonomy class.
Table 2. Excerpt from the Biomimicry Taxonomy

A taxonomy is generally an information structure used to classify instances. In a typical taxonomy, instances can be positioned into mutually exclusive, unambiguous categories, for example, the taxonomy of Linnaeus (Reference Linnaeus1767). However, the Biomimicry Taxonomy allows the classification of a single strategy into multiple categories. Take, for example, the harlequin beetle: an organism that uses its strong, large mandibles to escape from the trees in which it is born by chewing through wood. This strategy is currently classified in the following two categories: “break down” / “physically break down” / “biotic materials” and “move or stay put” / “move” / “in solids.” Although one can argue about the word choice of taxonomy in the Biomimicry Taxonomy, the fact that biological strategies can be positioned into more than one category does not impede the functioning of the bioinspiration tool. In contradiction, this property allows the retrieval of a single biological strategy document for different relevant problem formulations or desired functions.
5. AUTOMATED CLASSIFICATION INTO THE BIOMIMICRY TAXONOMY
On September 18, 2012, the AskNature database contained 1531 unique strategy descriptions, with a total of 2826 classifications into the Biomimicry Taxonomy. Forty-seven percent of all strategies are classified into only one third-level Biomimicry Taxonomy class, 32% are classified twice, 13% three times, and 8% more than three times; the average number of classifications per unique biological strategy is 1.82. For the proposed automated classification approach outlined in this section, the 1531 reference documents, with their 2826 classifications, form the reference corpus. Before detailing the processing steps of the automated classification algorithm, in order to understand the choices that were made, a number of challenges of the classification task are the following:
• Challenge 1: The total number of reference documents (1531) and the total number of their classifications (2826) are low compared to the total number of Biomimicry Taxonomy classes (159). This makes training examples a scarce resource.
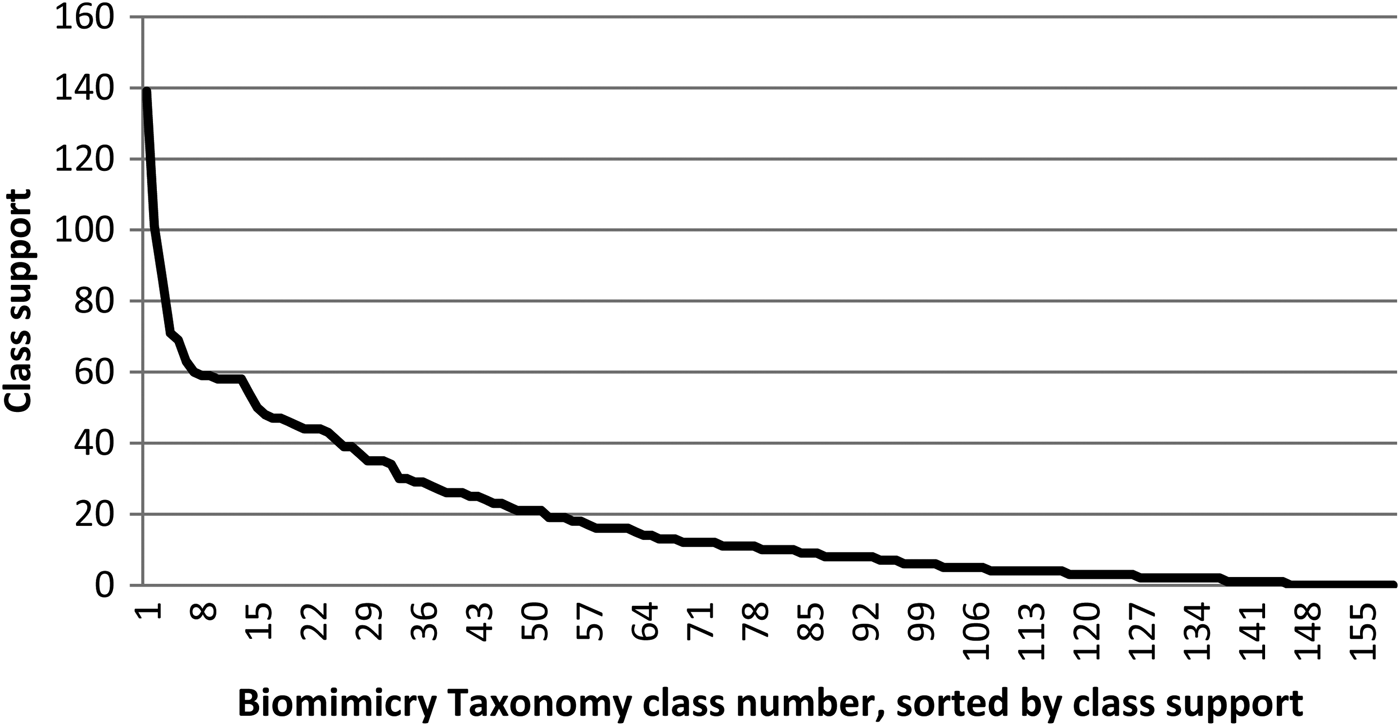
• Challenge 2: The number of reference documents per class, further referred to as class support, is not evenly distributed over these classes. Figure 1 shows the reference document support per Biomimicry Taxonomy class. Such an uneven distribution of reference documents encumbers training an automated classifier because some classes will not have enough reference documents for trustworthy decision making. There are 18 classes that, for the moment, have no reference strategies assigned to them.
• Challenge 3: The reference documents are short, with on average 435 words per document. Fewer terms in the reference documents entail fewer links to the sample documents. In contrast, a well-written short description of a biological strategy should contain a relatively large number of relevant document features for automated classification into the Biomimicry Taxonomy.
• Challenge 4: The Biomimicry Taxonomy classes are not mutually exclusive, as explained in Section 4. This introduces ambiguity in the classification task, which has on average 1.82 correct classifications per biological strategy.
• Challenge 5: Not all possible classifications for each unique strategy are exhaustively identified in the reference corpus. Although this has no consequences for the correct functioning of the bioinspiration tool, when reference documents are used as sample documents during the testing of the classification algorithm, this is likely to lead to performance underestimation.

Fig. 1. Distribution of the number of available reference documents (or support) per class; the class names of the 10 best-supported classes are provided in Table 4.
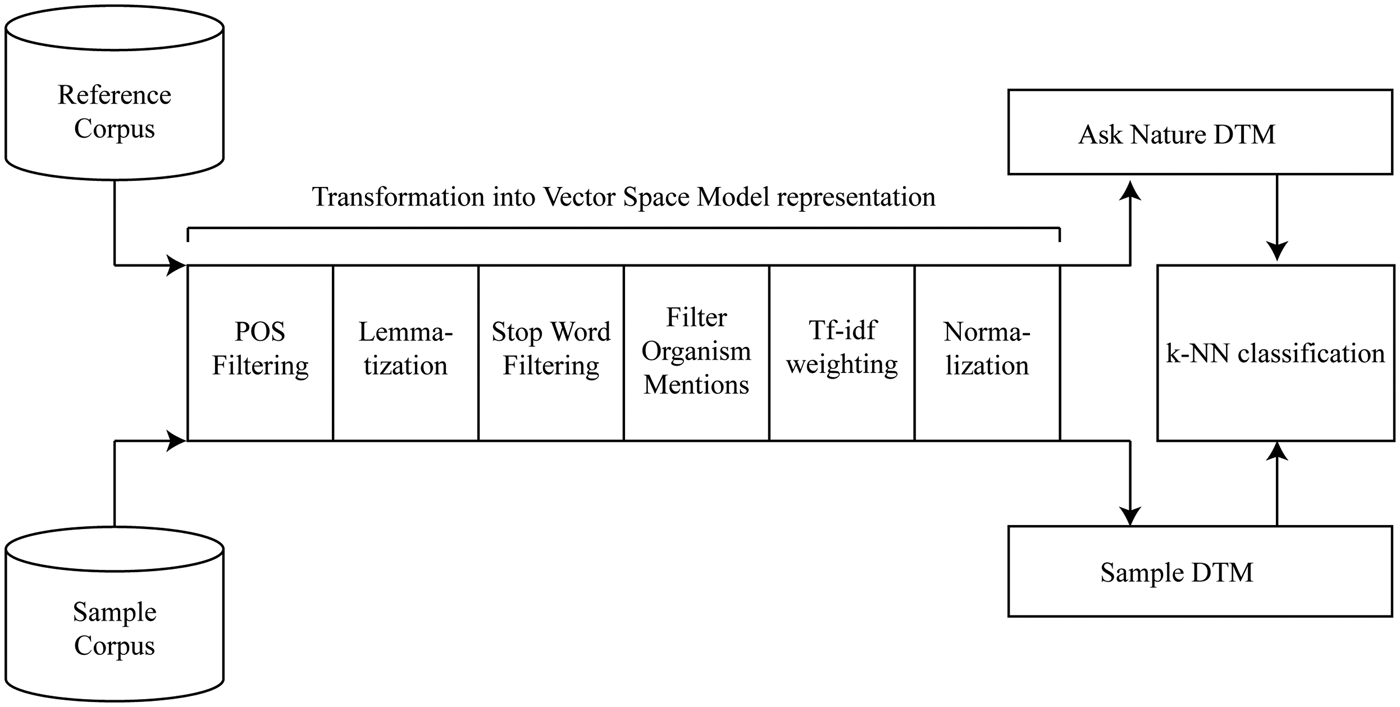
The answers to these challenges are detailed throughout the description of the proposed system, its testing, and discussion. Figure 2 provides an overview of the proposed approach. A number of preprocessing steps transform the reference and sample corpus into document-term matrices. The reference corpus contains the strategies of the AskNature database, and the sample corpus can contain any number of biological strategies in natural-language format. The document-term matrices of these corpora are their representations in the vector space model (Salton et al., Reference Salton, Wong and Yang1975). This algebraic model represents documents as vectors, where each dimension corresponds to a unique corpus word or feature and each feature value corresponds to the importance of the word in the document. In this vector space representation, mathematical operations between reference and sample document vectors enable the calculation of interdocument distances. For the illustrated algorithm, k-NN classification (Cover & Hart, Reference Cover and Hart1967) identifies a target classification for each sample document. The sections below detail the different steps of the proposed Biomimicry Taxonomy classification system.

Fig. 2. The algorithm for automated classification into the Biomimicry Taxonomy.
5.1. Reference and sample corpus
The reference corpus, depicted in Figure 2 as AskNature corpus, consists of all AskNature's strategies and their reference classifications as observed on September 18, 2012. The process of reference corpus building is illustrated in Appendix B with pseudocode. From AskNature's strategy pages the title, subtitle, summary, and excerpt are combined to form the strategy description. This results in a reference corpus with relatively short documents, as described in Challenge 3. Therefore, where possible, the extra sources, referred to on the strategy pages, are added to the reference documents. These extra sources are books, academic papers, and Internet articles. All books and a small number of academic papers and Internet articles listed as source for more than two different strategies are omitted as extra source because their content is too general. The remainder of the academic papers and Internet articles are retained as candidate extra sources. In this way, the 1531 reference documents have in total 1140 unique academic papers or Internet articles as possible extra sources, of which 886 were found to be retrievable. An algorithm leveraging the Google API retrieved 436 of these, 450 were added manually, and 254 were proved not retrievable.
The sample corpus can be any number and any type of textual description of specific biological strategies. Biological strategies are mainly documented in books, academic papers, and Internet articles, all of which can be integrated by the proposed system as the algorithm applies to documents in natural-language format. However, it is necessary that individual biological strategies are segmented in separate documents. Thus, biological books, such as Life: The Science of Biology (Purves et al., Reference Purves, Sadava, Orians and Heller2001), containing many different strategies in one document, should be subdivided with a topic identification algorithm (D'hondt et al., Reference D'hondt, Verhaegen, Vertommen, Cattrysse and Duflou2011) into smaller topic-specific documents before classification is attempted. Academic papers and Internet articles are typically directed toward a single topic and require no segmentation. Academic papers can be obtained from relevant journals, and both academic papers and Internet articles can, for instance, be gathered by a thereto-trained focused webcrawler (Vandevenne et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2011). As detailed in Section 6, in order to avoid possibly subjective expert evaluations, different small sample corpora are isolated from the reference corpus for validation purposes.
5.2. Transformation into vector space model representations
All preprocessing steps, transforming the reference and sample corpus into vector space model representations, are identical for both corpora and illustrated in Appendix B with pseudocode. First, part of speech (POS) tagging (Charniak, Reference Charniak1997) is performed with a standard TnT tagger (Brants, Reference Brants2000), and only the verbs, adverbs, adjectives, and nouns are retained for further processing. Second, this POS information guides WordNet-based lemmatization (Stark & Riesenfeld, Reference Stark and Riesenfeld1998) for the remaining corpus terms. Lemmatization eliminates all terms that are not inflections of WordNet lemmas. For instance, lemmatization allows the linking of one document mentioning biting to another document mentioning bites through association of both documents with the lemma bite. Lemmatization is an important preprocessing step because it identifies additional document linkages between the sample documents and the short reference documents (Challenge 3). In the third filtering step, stop words (Fox, Reference Fox1989) are removed, because they do not represent relevant document content. Fourth, the occurrences of organism names in the texts are also filtered to avoid the situation where many interdocument links would be caused by irrelevant organism names instead of terms related to the described biological strategy. Filtering organism names is comparable with filtering words related to products in Verhaegen et al. (Reference Verhaegen, D'hondt, Vandevenne, Dewulf and Duflou2011) because in both approaches interdocument links are removed to bring out structure relevant for design by analogy. Omitting organism name filtering can cause, for example, a strong but undesirable link between a sample document discussing the strong turtle bites and a reference document detailing turtle shields. Organism name detection is performed by LINNAEUS (Gerner et al., Reference Gerner, Nenadic and Bergman2010), an open-source species name identification system. Its database, containing only names at sthe pecies level, is expanded to include all scientific and common organism names of the National Center for Biotechnology Information taxonomy. Because biological strategies often contain mentions of ranks higher than the species level, all 26 biological ranks are included. The above preprocessing steps, converting text documents to document vectors, are illustrated in Appendix C with a running example. The two final preprocessing steps are term frequency-inverse document frequency weighting and normalization. Term frequency-inverse document frequency weighting (Salton & Buckley, Reference Salton and Buckley1988) gives more importance to terms occurring in a limited set of documents and less importance to terms occurring in many documents of the corpus. Normalization, finally, compensates for the differences in document size.
5.3. k-NN classification into the Biomimicry Taxonomy
The k-NN is a machine learning approach that takes the k-nearest neighboring reference documents to a sample document, and, to compose a hypothesis, performs a majority vote on those reference document classifications (Cover & Hart, Reference Cover and Hart1967). The k-NN is a type of instance-based learning, meaning that instead of making a generalized classification model, a classification hypothesis is directly constructed from the reference corpus documents. For the specific classification task at hand, the lack of model building is an especially interesting property. As the reference database slowly grows, that is, strategies are currently manually added by a rate of one every couple of days, new documents can be added to the reference set without the need to recalculate the model. Training should be seen as merely storing the new feature vectors with their reference classifications. For the proposed system, the decision of which k reference documents are the nearest neighbors of a sample document is based on the well-known cosine similarity measure between their document vector representations (Baeza-Yates & Ribeiro-Neto, Reference Baeza-Yates and Ribeiro-Neto2011). The more similar reference and sample documents are, the smaller the angle is between their feature vectors, and the more their cosine similarity approaches 1. Completely dissimilar document vectors are orthogonal; hence their cosine similarity is 0. The parameter k, which represents the number of reference documents that take part in the majority vote, currently is set to 50. However, the value of this parameter should be reevaluated when the reference corpus support increases significantly. The process of k-NN classification is illustrated in Appendix D with pseudocode.
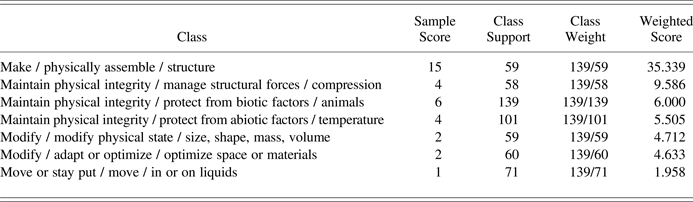
Performing a majority vote on the classifications of the k-nearest neighboring reference documents results in an ordered list of candidate Biomimicry Taxonomy classes, as illustrated for an example biological strategy document with the title “Fibers reinforce nests: wasps” in Table 3. The first two columns represent, respectively, the target classes and the number of reference documents in the k-nearest neighbors associated to these classes (depicted as “sample score”). In order to prevent the bias in corpus support, illustrated by Figure 1 and column 3 in Table 3, from weighing on the classification results, the majority vote is counterweighted accordingly with the weights shown in the fourth column. These are calculated for each class as the maximum support divided by the class's support. Given the class support distribution illustrated by Figure 1, it can be easily seen that such weighting currently prevents the classes with very low support from taking part in the majority vote. For example, in the most extreme case, a class with only one reference document would receive a weight equal to the number of reference documents in the best-supported class and, hence, would dominate voting results. In the ideal case, when all target classes contain a sufficiently high and equal number of reference documents, weighting will be omitted and all classes will take part in the majority vote. However, considering the current status of the reference corpus (Challenges 1 and 2), for the time being, classification is limited to the top 10 Biomimicry Taxonomy classes. Applying this class filter to the full list of candidate classifications linked to the example document's k nearest neighbour reference documents results in the short list of candidate classes shown in Table 3. The highest weighted score is taken as classification for the sample document. In this example, 15 out of the 50 nearest neighbouring reference documents have the classification make / physically assemble / structure, which agrees with the reference classification of the example biological strategy.
Table 3. Example of an ordered list of candidate Biomimicry Taxonomy classes

6. RESULTS
The proposed Biomimicry Taxonomy classification algorithm is tested to confirm that it is able to classify new biological strategies. Random selections of strategies, 30 for each test, are isolated from the reference corpus to build the sample corpus, for the following reasons. First, in this way, there is sufficient confidence that the sample documents are relevant biological strategies that deserve to be positioned into the Biomimicry Taxonomy. Second, these sample documents each have one or more reference classifications, validated by AskNature, which are used as a golden key. Third, one can assume that most strategies in the AskNature reference corpus contribute something new to the knowledgebase; or in other words, that there are few reference strategies describing the same biological strategy or phenomenon. This last test set property ensures that the sample documents actually represent new biological strategies, instead of adding similar or identical sample strategies to the reference corpus.
As explained in the previous section, with the current status of the reference corpus (see Challenges 1 and 2), one cannot evaluate automated classification for all Biomimicry Taxonomy classes. Hence, automated classification is currently evaluated for the 10 best-supported classes, listed in Table 4. From these classes, 30 random reference strategies are selected for each test. These randomly selected strategies and all their Biomimicry Taxonomy classifications are removed from the reference corpus before applying the procedure depicted in Figure 2. The main reason for testing in smaller batches of 30 is to avoid significant further reduction of the reference corpus class support, which is likely to cause underestimation of performance.
Table 4. Top 10 supported Biomimicry Taxonomy classes

Note: GSDR, get, store, distribute resources.
An example of the detailed results of the first test run is provided in Appendix A. For this sample corpus, 18 out of the 30 sample documents are given a classification hypothesis that complies with AskNature's reference classification. Table 5 summarizes the results of eight test runs of 30 randomly selected sample documents. Averaging these results amounts to a classification precision of 61.25%. However, this performance number is only aimed at illustrating the potential of the approach, not estimating its final performance on all categories once the Biomimicry Taxonomy classes are adequately supported by reference documents. In addition, because not all possible classifications in the reference corpus are exhaustively identified (Challenge 5), it is likely that the real number of correct classifications is higher than we can provide in Table 5. For example, the strategy named “Secondhand weapons protect from predators: sea slug” has only one reference classification: “Maintain physical integrity” / “Protect from biotic factors” / “Animals.” The algorithm's first hypothesis for this sample document is “Get, store, or distribute resources” / “Capture, absorb, or filter” / “Organisms”; therefore, this is counted as an incorrect classification. However, the sea slug eats its way into jellyfish, and the stinging cells of the latter migrate unchanged to the tentacles on the back of the former where they give the same protection as for their previous owner who developed them (Attenborough, Reference Attenborough1979). The slug thus absorbs parts of its victim to build and extend its own protection. The algorithm picked up on this and proposed a viable candidate classification currently not recognized as reference classification. A more straightforward example is the classification into “Move or stay put” / “move” / “in or on liquids” for the strategy with title “Pressure allows movement: echinoderms” and subtitle “Legs and tubes in echinoderms such as starfish allow movement and feeding by use of hydrostatic pressure.” Again, this classification is correct but not present in the reference corpus and thus counted as an incorrect classification of the algorithm in Table 5.
Table 5. Summary of test results, scores on 30 per test

In order to illustrate the potential of the algorithm to propose multiple correct classifications for a specific sample document (see Challenge 4), one has to take into account all reference documents for the majority vote instead of only those belonging to the 10 best-supported classes. In order to allow for this illustration, it must be noted that, as explained in Section 5, weighting needs to be omitted and the bias in reference corpus distribution is not counteracted. This, however, does not impede interesting results from coming forward. For example, Table 6 shows the top five proposed classifications for the strategy named “Bioluminescence protects from predation: dinoflagellates.” AskNature's reference database lists four possible classifications; all returned by the algorithm as first, second, fourth, and fifth guess. The third guess, in this handpicked example, Get, store, or distribute resources / Capture, absorb, or filter / Organisms, is also relevant once one understands the strategy in detail. When disturbed, dinoflagellates light up and create a glowing trail that leads predators higher up in the food chain to the attackers of the dinoflagellates (Haddock et al., Reference Haddock, Moline and Case2010); that is, the dinoflagellates use a kind of burglar alarm to capture their direct attackers, but they leave the capturing itself to a willing third party.
Table 6. Top five classifications given by the proposed algorithm for the example strategy: “Bioluminescence protects from predation: dinoflagellates”

7. DISCUSSION AND FUTURE WORK
There are three arguments that motivate the choice of scaling AskNature. First, the approach of AskNature is the only publicly available bioinspiration system with a continuously growing database that, at current, qualifies as an initial training set for scaling an existing BID ideation approach. Second, using the tool requires very little training, which adds to its appeal. By scaling AskNature's approach, the authors make no argument about the optimality of the specific adopted categorization. AskNature has updated the Biomimicry Taxonomy iteratively, and there is no indication that this process is final. Third, no research exists that identifies the most optimal tool for systematic BID. Therefore, the authors do not claim to have scaled the most optimal systematic BID approach, but to be the first to demonstrate the feasibility of scaling any BID approach, that is, AskNature, the most popular one. The tests in this paper are performed on the Biomimicry Taxonomy as observed on September 18, 2012, and the applied classification algorithm is independent of possible future changes to this taxonomy.
For testing the proposed approach, the classifications provided by AskNature are used as a golden key for the validation tests (see motivation in the previous section). As this database is collaboratively expanded, which is a human operation, it is possible that some error is introduced in the training and test sets. During the interactions with this reference database while testing, the authors have not encountered misclassifications of AskNature strategy documents. Therefore, in the context of illustrating the feasibility of the proposed approach, the reference classifications are taken as a golden key. However, as part of future work, checking the reference database against human raters would be valuable to verify the high level of trust AskNature's reference classifications currently enjoy. Besides quantifying the fraction of potential misclassifications, such validation could also quantify the observation that for some reference documents not all relevant reference classes are exhaustively identified in the reference corpus (see Section 5, Challenge 5).
Four typical steps of the systematic BID process are problem formulation, solution search, filter and analysis of alternatives, and knowledge transfer (Vandevenne et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2011). By scaling AskNature, the authors have addressed an important bottleneck in the second step: solution search. However, choosing AskNature in the search phase does not exclude other approaches from being integrated in the scaled systematic BID process to come to a hybrid approach. Because all mentioned related research approaches described in Section 3 have a functional component in their problem formulation, in the first step, restating the problem with other guidelines than those of AskNature can inspire us to look at the problem differently and result in new entry points into the Biomimicry Taxonomy. Furthermore, the three model-based systematic BID approaches can support both the analysis of a small number of alternative strategies (Step 3) and the formulation of knowledge transfer (Step 4). When composing any of these models for the low number of alternative biological strategies, the designer is forced to look at the system from different perspectives (e.g., function, behavior, and structure). This helps to form a more complete understanding of biological systems under focus. If there are problems understanding the retrieved biological solutions, the authors envisage a need to contact domain-specific biologists in these two final steps. Instead of asking biologists to identify what is interesting in nature to the design problem at hand (search phase), their role is shifted to the analysis and knowledge transfer phases. Besides a wider search, not limited to the personal knowledge of one or a couple of biologists, this allows to us contact the most relevant biologist(s) for the specific design problem.
The more reference documents will support the classes of the Biomimicry Taxonomy, the more relevant biological strategies will be able to take part in the majority vote. Therefore, taking into account the current scarcity of training examples (Challenge 1), it is reasonably expected that the performance of the system will increase further when the reference corpus grows. In addition, the randomly drawn sample documents from the reference corpus are much shorter documents (on average 435 words per document) than typical biological papers or online articles (thousands of words) that require automated classification. Because shorter documents are likely to have fewer interesting features to form relevant interdocument links, this makes the sample corpus extra challenging. Hence, the reported performance of the outlined approach should be interpreted as a proof of concept that illustrates the potential of the approach. Because any classifier is bound to make at least some misclassifications, the authors envisage a user interface element allowing users to signal the system that a certain strategy is positioned incorrectly. When sufficient human raters agree about such a classification adjustment, the misclassification of the sample document can be corrected and the document can be even moved to the reference corpus.
Every step in the classification procedure runs completely autonomously (hence, the proposed classification approach scales for leveraging large biological databases). Nevertheless, there are two notes to be made in the context of scalability. First, the reference corpus is composed manually by AskNature's online community, with steady but relatively slow growth. In order to realize sufficient support for all classes of the Biomimicry Taxonomy, the community should be steered toward adding strategies for those categories where necessary, instead of directing attention to categories that, from the point of building a reference corpus, are not a priority. Second, by anticipating a small user interface element allowing people looking for inspiration to signal a strategy's potential misclassification, again a light form of crowd sourcing is proposed to further boost the system's precision. However, this adds to the current state of the art because the required task is in the order of a single mouse click for a relatively small number of biological strategies, and once the reference corpus is adequately built, this will be the only manual interaction in support of the scaled ideation system.
One way of obtaining a sample corpus is training a webcrawler to collect biological strategies from the Internet (Vandevenne, Caicedo, et al., Reference Vandevenne, Caicedo, Verhaegen, Dewulf and Duflou2012). Such strategy documents can be fed to the presented classifier as sample corpus, which was previously done with an early version of the classification algorithm (Vandevenne, Verhaegen, et al., Reference Vandevenne, Verhaegen, Dewulf and Duflou2012). In these tests, precision was boosted to 90% by introducing a cutoff score on the sample document classification results. This, however, entailed that most sample documents did not have a target classification that met the cutoff score and were discarded. To avoid such loss of sample documents, the presented algorithm in Section 5 does not integrate a cutoff score. However, in the context of a webcrawling corpus, that in theory could become very large, adopting a cutoff score to boost precision can be argued for.
Although Section 6 presented tests that indicate it is feasible to scale AskNature's database expansion with automated classification, doing so will introduce a new challenge: avoiding information overload. When a designer selects a Biomimicry Taxonomy functional class to retrieve biological strategies relevant to his or her problem, presenting very large lists of strategies should be avoided. Therefore, the authors anticipate the need for a hierarchical representation of the results based on similarities between the strategy descriptions. To implement this user interface functionality, the suitability of standard document clustering techniques will be explored, to compose an extra information level for assisting the designer with the efficient identification of relevant strategies. In addition, when biological strategies in the form of academic papers are returned to the person looking for bioinspiration, the size of these documents (thousands of words per strategy) necessitates highlighting specific paragraphs, sentences, or words. To facilitate analogical transfer, efforts should be made to assist in the identification of terms relevant for structural mapping (Gentner & Markman, Reference Gentner and Markman1997) between the source and target domains. Highlighting the most important words that caused the classification of the retrieved biological strategies into the chosen functional Biomimicry Taxonomy class could be a good start. Furthermore, to accentuate words representing causally related functions, a recently developed extraction algorithm (Cheong & Shu, Reference Cheong and Shu2009, Reference Cheong and Shu2012) could be integrated to further support the abstraction of biological strategies to facilitate cross-domain knowledge transfer.
8. CONCLUSION
This paper proposes an algorithm that allows the fully automated integration of any number of biological strategies into the bioideation tool of AskNature. In this way, an answer is given to the scalability challenge, posed by the ever growing body of knowledge about nature, that current ideation tools typically struggle with. Tests are presented that validate the potential of the approach for those Biomimicry Taxonomy functional classes that currently hold sufficient reference strategy support. Reported performance is encouraging, although, for the moment, limited by the current status of the reference corpus. After this proof of concept, the logical next step is to further expand the reference corpus for the remaining Biomimicry Taxonomy classes in order to fully scale the approach.
Dennis Vandevenne has been a Researcher at Katholieke Universiteit (KU) Leuven since 2006. Until 2009 he performed research on identity management and biometrics in the Department of Computer Security and Industrial Cryptography. He currently performs research on methods and algorithms for BID at the Centre for Industrial Management. Dennis holds three information and communications technology (ICT) related masters: electronics-ICT, artificial intelligence engineering and computer science, and industrial management-ICT.
Paul-Armand Verhaegen founded and sold Stocks, a company specialized in printer supplies. He worked as an external consultant for 3E on green energy certificates and as a consultant at Bureau van Dijk Management Consultants. Paul-Armand was employed at Vrije Universiteit Brussel, Erasmushogeschool Brussel, and KU Leuven as a Research Assistant. Dr. Verhaegen has a master's degree in applied science and engineering, with a specialization in electrotechnical and computer science; a graduate degree in the complementary studies in business administration; an MBA from Vrije Universiteit Brussel; and a PhD in engineering from KU Leuven. He is currently pursuing a PhD in the domain of systematic innovation.
Simon Dewulf is founder of CREAX. CREAX develops the www.creationsuite.com, a web-based open innovation interface. CREAX Creation Suite is used by companies such as Dow Corning, SKF, P&G, Philips, Kraft, L'Oreal, Johnson & Johnson, and GSK to provide direct access to out of domain knowledge for technology transfer and open innovation opportunities.
Joost R. Duflou is a Professor in the Department of Mechanical Engineering at KU Leuven. He has master degrees in architectural and electromechanical engineering and a PhD in engineering from KU Leuven. He is a member of CIRP and has been published in over 200 international publications. His principal research activities are in the field of design support methods and methodologies, with special attention for systematic innovation, ecodesign, and life cycle engineering.
APPENDIX A
Classifications of the first test of 30 random reference documents from the 10 best supported classes

APPENDIX B
The pseudocode below explains the logic of the implementation described in section 5.1: reference and sample corpus creation. The actual implementation is optimized for performance.

APPENDIX C
The pseudocode below explains the logic of the implementation described in section 5.2: Transformation into Vector Space Model representations. The actual implementation is optimized for performance.

APPENDIX D
The pseudocode below explains the logic of the implementation described in section 5.3: k-NN classification into the Biomimicry Taxonomy classes. The actual implementation is optimized for performance.

APPENDIX E
The following is a short running example to illustrate the conversion of plain text to a document vector, as described in Section 5.2 and Appendix C.
Sample text, a short biological text: Geckos use opposing feet and toes while inverted, possibly to maintain shear forces that prevent detachment of setae or peeling of toes. If detachment occurs by macroscale peeling of toes, the peel angle should monotonically decrease with applied force.
Tokenization, POS tagging, and filtering non-(verb, adjective, noun, adverb) terms: [noun] geckos [verb] use [verb] opposing [noun] feet [noun] toes [adjective] inverted [adverb] possibly [verb] maintain [adjective] shear [noun] forces [verb] prevent [noun] detachment [noun] setae [verb] peeling [noun] toes [noun] detachment [verb] occurs [noun] macroscale [verb] peeling [noun] toes [noun] peel [noun] angle [adverb] monotonically [verb] decrease [adjective] applied [noun] force
Lemmatization, replace terms by their lemmas or removes terms if they are not present in the WordNet thesaurus with the POS tag: [noun] gecko [verb] use [verb] oppose [noun] foot [noun] toe [adjective] inverted [adverb] possibly [verb] maintain [noun] force [verb] prevent [noun] detachment [noun] seta [verb] peel [noun] toe [noun] detachment [verb] occur [verb] peel [noun] toe [noun] peel [noun] angle [verb] decrease [adjective] applied [noun] force
Mention and stop word detection: [noun, mention] gecko [verb, stop word] use [verb] oppose [noun] foot [noun] toe [adjective] inverted [adverb] possibly [verb] maintain [noun] force [verb] prevent [noun] detachment [noun] seta [verb] peel [noun] toe [noun] detachment [verb] occur [verb] peel [noun] toe [noun] peel [noun] angle [verb] decrease [adjective] applied [noun] force
Mention and stop word filtering: [verb] oppose [noun] foot [noun] toe [adjective] inverted [adverb] possibly [verb] maintain [noun] force [verb] prevent [noun] detachment [noun] seta [verb] peel [noun] toe [noun] detachment [verb] occur [verb] peel [noun] toe [noun] peel [noun] angle [verb] decrease [adjective] applied [noun] force
Document vector representation: