INTRODUCTION
Verocytotoxin-producing Escherichia coli (VTEC), especially those of serotype O157:H7, are causally related to diarrhoea, haemorrhagic colitis (HC), and the potentially lethal haemolytic–uremic syndrome (HUS) in humans [Reference Tesh and O'Brien1–Reference O'Brien, Kaper, Kaper and O'Brien3]. Humans most often become infected with VTEC through consumption of contaminated foods (meat, milk, or raw vegetables) or by direct transmission from patients or infected animals [Reference Dev, Main and Gould4–Reference Heuvelink9]. The majority of outbreaks are related to cattle or products of bovine origin [Reference Griffin and Tauxe10, Reference Meng, Doyle, Kaper and O'Brien11]. Epidemiological studies have identified cattle as the main host for E. coli O157 and other VTEC [Reference Armstrong, Hollingswoth and Morris7, Reference Chapman12–Reference Schouten16].
The presence of O157 VTEC in cattle and their environment has been investigated [Reference Schouten14–Reference Garber19], and several studies have shown a seasonal effect in prevalence of O157 VTEC: the shedding season appears to be summer and early autumn. Duration and magnitude of E. coli O157:H7 shedding in cattle faeces and localization of the bacterium in the gastrointestinal tract of experimentally inoculated cattle also have been studied [Reference Brown20–Reference Zhao23]. Despite these studies, little is known about the dynamics of O157 VTEC within cattle farms, although transmission within a population has been quantified [Reference Laegreid and Keen24–Reference Matthews26]. Transmission can be assessed quantitatively by using experimental or field data and can be expressed by the basic reproduction ratio (R 0), which is defined as the average number of secondary cases caused by one typical infectious individual during its entire infectious period in a fully susceptible population. If R 0>1, each infected animal will infect on average one or more susceptibles, which might lead to a major outbreak [Reference Kermack and McKendrick27, Reference Jong and Kimman28].
An important question regarding transmission dynamics of O157 VTEC is whether and how previously positive animals or herds become infectious in the next shedding season, after a period of non-shedding. First, stress or dietary changes, specifically in the summer months, might trigger shedding of potentially latent O157 VTEC carriers. Second, in addition, water, soil, and manure can be long-term reservoirs for E. coli O157 [Reference Faith29] and re-infection of animals may occur, but no studies have been performed to assess pastures for infectivity of soil and manure.
The aim of this paper is to compute the reproduction ratio (R 0) for O157 VTEC from a transmission experiment with dairy calves and from a longitudinally sampled dairy herd known to be positive for O157 VTEC. Additionally, we investigated the contribution of animals and pastures (soil and manure), previously determined as being positive, to cause infection in the next season.
METHODS
Transmission experiment
In September 2000, 20 Holstein Friesian heifer calves 12–15 weeks old were purchased from a single farm. Each calf tested negative for O157 VTEC three times during the 3 weeks before arriving at the experimental facilities, and were considered susceptible. Calves were randomly assigned to one of the two groups (groups 1 and 2) of 10 animals each: each group was randomly assigned to one of two paddocks, 2000 m2 each. A pool of multiple samples of the soil of these paddocks was tested three times with a 1-week interval, and O157 VTEC was not detected.
After an adaptation period of 14 days, during which calves were tested twice for O157 VTEC, five randomly selected calves of each group were housed in one of two climate-controlled units with group housing, and were inoculated orally with 109 colony-forming units (c.f.u.) of a doxytetracycline-resistant strain of E. coli O157, containing VTI, VTII and eae-encoding genes (strain 20G8), which were isolated from cattle faeces in a study by Heuvelink et al. [Reference Heuvelink13]. The units had separate floor drains, and faeces were removed daily. When inoculated calves were considered infectious (i.e. colonized: microbiologically positive, followed by shedding for three consecutive days), they were reunited with the group of susceptible contact calves they originated from. Calves were clinically normal at the time of inoculation and at the time of rejoining their group.
Calves were fed concentrates and grass-pelleted meals in an amount appropriate for their age, to make sure that they had sufficient roughage (on pasture and in the climate-controlled units). Water was provided ad libitum. Concentrates and grass-pelleted meals were irradiated (γ-irradiation: 9 kgrey) to prevent foodborne infections.
From day 0 (day of inoculation September 2000), both inoculated and susceptible contact calves were examined daily for 14 days for clinical abnormalities, such as diarrhoea, pyrexia, and anorexia. Rectal faecal samples were taken daily and cultured to detect O157 VTEC.
On day 40, 14 of the 20 calves were euthanized with sodium pentobarbital and examined by necroscopy and histologically for presence of attaching-and-effacing lesions in the gastrointestinal tract. The remaining six (contact-infected) calves that almost continuously shed O157 VTEC during the experiment were housed indoors and tied individually, so that the probability of infecting each other was minimized. After day 40 (October 2000), faecal and blood samples were collected three times a week for 8 weeks, then twice a week for 4 weeks. From then on, calves were tested weekly for another 6 months, until June 2001.
The Ethical Committee for Experimentation with Animals (Lelystad, The Netherlands) approved the experimental protocol.
Previously infected calves and pastures
A second experiment was performed to investigate the possibility of formerly contact-infected calves and pastures functioning as reservoirs for O157 VTEC between shedding seasons. Three pastures were used that lay fallow during winter and from which one grass cut was harvested in spring.
In June 2001, six previously contact-infected heifer calves of the transmission experiment that tested negative for O157 VTEC since day 81 were placed in a pasture that had not been grazed on by ruminants for at least 1 year, and that had tested negative for O157 VTEC four times during April and May 2001 (‘clean pasture’). At each sampling round, four pooled soil samples, each consisting of 10 random samples of about 20 g taken in a bag at appointed sites in the paddocks (0–5 cm deep), were analysed for O157 VTEC.
Further, in June 2001, 10 purchased bull calves that tested negative for O157 VTEC three times were divided into two groups of five and randomly assigned to one of two pastures that were contaminated with O157 VTEC during the transmission experiment of the previous season.
During these two experiments, faecal and blood samples were collected weekly from all animals for 14 successive weeks. In addition, soil samples were taken twice a month, from each of three pastures as described above.
Field transmission study
A dairy farm situated in the centre of The Netherlands was selected from a group of dairy farms that tested positive for O157 VTEC in a monitoring programme. The longitudinal study, from this point referred to as the ‘field study’, started July 1999 and ended November 2000. During this period, animals were sampled 14 times, at 4- to 10-week intervals. For details of animal population, housing, farm management, and sampling strategy see Schouten et al. [Reference Schouten16]. Herd size averaged 75 Holstein Friesian cows, of which 20% on average were non-lactating at each sampling. Non-lactating cows were housed separately from lactating cows. Faecal samples were collected from all cows by rectal palpation and placed in plastic bags. Because the O157 VTEC status of dairy cows on this farm was assessed repeatedly, transmission between cows could be quantified.
Sample processing and analysis
Faecal samples were transported in cool boxes and analysed within 24 h of sampling by isolation methods as described in Schouten et al. [Reference Schouten14]. Isolation per 10 g faeces was performed using enrichment in mTSB+A, subsequent immunomagnetic separation (IMS), incubation on sorbitol MacConkey agar (SMAC) (Oxoid, Basingstoke, Hampshire, UK) with cefixime and tellurite (CT-SMAC), and screening for sorbitol-negative colonies. Colonies were incubated on both SMAC supplemented with 4-methylbelliferyl-β-d-glucuronide (MUG; Sigma Chemical Co., St Louis, MO, USA) and eosin methylene blue agar (EMB; Oxoid). Suspected E. coli O157 colonies were tested by agglutination to ascertain authenticity. Within the transmission experiment, a sample was considered positive, based on the outcome of the agglutination test. Isolates of the field study that were confirmed by agglutination were also serotyped. A sample was considered positive when serotyping identified the isolate as E. coli O157. Isolates were subsequently screened by polymerase chain reaction for possession of genes encoding for the most common verocytotoxins (VTI and VTII) and the eae gene [Reference Schouten16].
Statistical analysis
For proper analysis of the data, it is essential to determine the ‘infection status’ – susceptible (S) or infectious (I) – of the individual animals at each sampling in each of the studies. Although infection implies a disease process associated with O157 colonization, which is not the case in cattle, ‘infectious’ is the usual term in transmission models to indicate the shedding state of animals that are capable of infecting (colonizing) susceptible animals.
Different assumptions regarding infection status were made. In the field study, the O157 VTEC status for each lactating cow was determined for each of the 14 samplings, assuming a positive-tested animal to be infectious (I) and a negative-tested animal to be susceptible (S). For the transmission experiment, the status for each calf was determined for each sampling; a calf was defined to be infectious (I) when found positive at three consecutive samplings or to be susceptible (S) when found negative at three consecutive samplings. These assumptions are based on the possible occurrence of intermittent excretion of E. coli O157-infected animals [Reference Brown20, Reference Zhao23, Reference Faith29] and that daily sampling was performed in the transmission experiment, whereas in the study on the dairy farm sampling took place about every 4–6 weeks.
A series of events, i.e. infectious contacts or incidents of new infections (cases) at consecutive samplings, can be considered a stochastic process. It was assumed that infectious animals stop shedding after a while and become susceptible again, meaning that the infection (colonization) gives no short-term immunity. Therefore, a susceptible–infectious–susceptible model (SIS model) was used to describe the transmission of O157 VTEC, both in the transmission experiment and the field study. The SIS model can be represented as:
In a population with size N (=S+I), the number of animals that are susceptible and become infectious per time interval Δt, depends directly on the transmission parameter β, the number of susceptible (S) and the number of infectious (I) animals. Infectious animals become susceptible again at a rate α. This implies that the mean length of the infectious period is 1/α. R 0 is then defined as β/α [Reference Jong and Kimman28].
The transmission parameter β can be estimated using a function of I, S, C, N, and Δt, defining the stochastic process based on a binomial distribution. For this, we assumed that all animals were randomly in contact, susceptible and infectious animals were homogeneous groups, infection rate was constant during the whole infectious period, and duration of the infectious period was exponentially distributed.
The number of new cases (C) at the end of each time interval (Δt) can be described by:
where S is number of susceptible animals (S(t)) at the start of the interval, I is average number of infectious animals (I(t)) during the interval, N is population size (=S+I for dairy herd;=10 for transmission experiment), and Δt is the sampling interval.
Taking the log of equation (1) results in
Data were statistically analysed using generalized linear models (GLM [Reference Becker30]). Applying GLM, the whole course of the infection chain is used to estimate the transmission parameter β. General linear regression (Stata® version 8; StataCorp, College Station, TX, USA) with a complementary log-log link function and log(I/N∗Δt) as an offset, was used, and S gives the number of trials for the binomial distribution.
The estimated parameter is log(β); exponentiation gives β. If the length of the total infectious period for O157 VTEC is known, R 0 can be calculated by multiplying the infection rate (β) with the length of the infectious period (1/α).
RESULTS
Descriptive results
Transmission experiment
No clinical disease was observed during the first days following inoculation with 109 c.f.u. E. coli O157:H7. Each of the ten inoculated animals shed the bacterium in their faeces for more than 3 days, starting day 1 post-inoculation (p.i.). From the day they started shedding, they were considered infectious. Because of the duration of the applied test (2 days in laboratory), infectious animals could not be reunited with contact animals until day 5 p.i. Contact calves tested positive 2 days after being reunited (day 7 p.i.) in group 1, and 1 day after being reunited (day 6 p.i.) in group 2. All contact animals tested positive within 4 days (day 9 p.i.) for group 1 and within 6 days (day 11 p.i), for group 2. In group 1, four of five inoculated animals tested positive at each subsequent sampling, except for the last sampling (day 39 p.i). In group 2, inoculated animals shed more intermittently; every inoculated animal tested negative in at least one sample. One calf tested positive for two consecutive samplings, within a range of negative samplings (−−++−−). This animal was classified as negative or susceptible. Other animals were either positive or negative for more than three consecutive samplings. The number of calves testing positive and negative at each sampling in the two groups is given in Table 1.
Table 1. Number of O157 VTEC-positive inoculated and contact calves by sampling day (days p.i.) and input for the transmission model for groups 1 and 2 of the experiment. At day 5, inoculated calves were joined with the susceptible calves
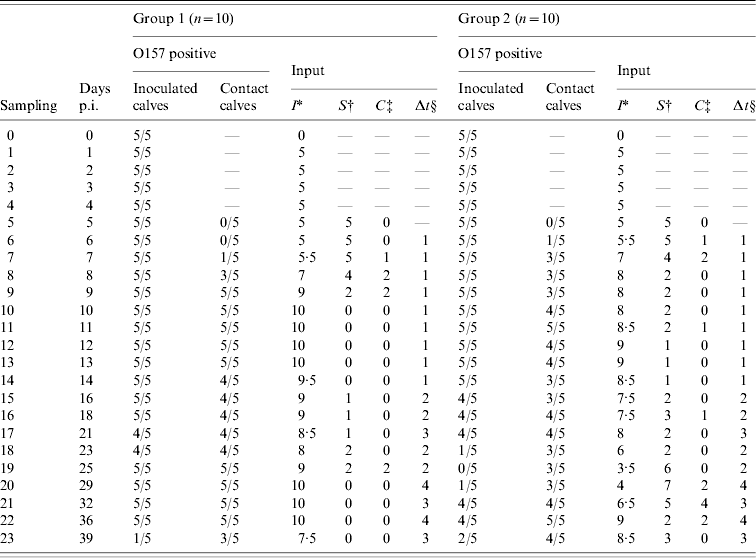
* Average number of infectious calves during the time interval preceding sampling. New cases were assumed to be infected halfway during the time interval on average.
† Number of susceptible calves at the beginning of the time interval preceding sampling.
‡ Number of new cases per interval of sampling.
§ Days between samplings.
At the end of the experiment (day 40), necropsy and histological examination of the rumen, reticulum, omasum, caecum, colon, ileum, and duodenum of the 14 calves (10 inoculated, four contact) did not show any attaching-and-effacing lesions.
On day 40, the six contact-infected calves most continuously shedding, with a positive culture outcome at day 36 p.i., were housed in a tie stall. In subsequent weeks, the number of calves shedding O157 VTEC declined. Up to day 81, four of the six calves were shedding intermittently, with three or fewer animals shedding at the same sampling. After day 81 p.i. each animal tested negative for O157 VTEC until the last day of sampling (day 100; Fig.).
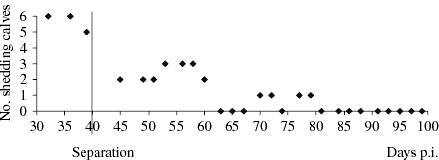
Fig. Number of shedders of six contact-infected calves by days post inoculation (p.i.) before and after separation of the group and individual tied housing.
Previously infected calves and pastures
O157 VTEC was not detected in the six previously infected animals while grazing on the ‘clean’ pasture. In addition, the 10 new susceptible calves on the previously infected pastures of the transmission experiment did not start shedding. O157 VTEC was not detected in any of the soil samples.
Field study
Detailed descriptive statistics of this study can be found in Schouten et al. [Reference Schouten16]. For each sampling, 0–29·5% of the cattle tested positive for E. coli O157, with prevalences of 0% during winter. Nine cows tested positive more than once, but all in consecutive samplings. At the end of the sampling period, O157 VTEC was isolated at least once from 41·4% of the cows in the herd.
Estimation of O157 VTEC transmission by statistical modelling
For the transmission experiment and the field study, numbers of susceptible (S), infectious (I), and new cases (C) per time interval were counted (Tables 1 and 2) and used as model input.
Table 2. Number of sampled and O157 VTEC-positive dairy cows, and input for the transmission model for the dairy cow population by sampling moment in the field study
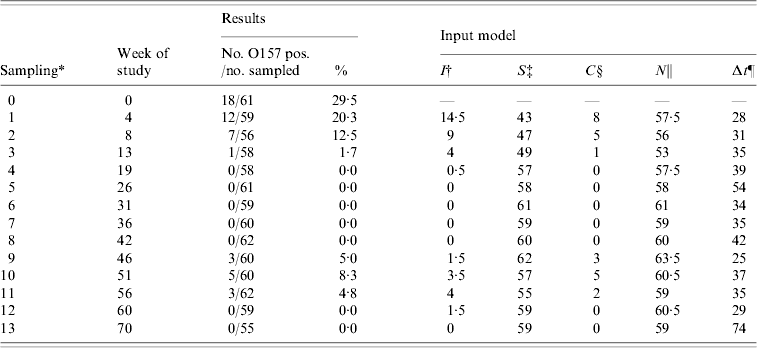
* Samplings 1, 2, 3, 4, 9, 10, 11, 12, and 13 were in the shedding season (5, 6, 7, 8 were December, January, February, March).
† Average number of infectious cows during the time interval preceding sampling. New cases were assumed to be infected halfway during the interval on average.
‡ Number of susceptible cows at the beginning of the time interval preceding sampling.
§ Number of new cases per interval of sampling.
ǁ N=I+S per time interval.
¶ Days between samplings.
To calculate R 0, the calculated values were multiplied by 28 days, assuming this to be the length of the infectious period. This length was based on the observed length of shedding in the experiment, the interval of sampling in the field study (which was at least 1 month) and the literature [Reference Brown20, Reference Cray and Moon21], and used only to give an indication of the magnitude of R 0. With this assumption, the R 0 value for group 1 was 10·3 (95% CI 5·0–21·9), for group 2 R 0=6·4 (95% CI 3·6–10·9), and overall R 0=7·3 (95% CI 3·9–11·5) (Table 3). Assuming that a positive calf is infectious immediately and that it is susceptible immediately when testing negative, the estimate for group 2 would increase to 7·0 (95% CI 4·2–12·0) and the overall estimate would increase to 7·9 (95% CI 5·3–12·3). For group 1, no changes were observed.
Table 3. Transmission coefficients (β) and reproduction ratios (R0) with their 95% confidence intervals (CI) estimated from data of the transmission experiment and the field study
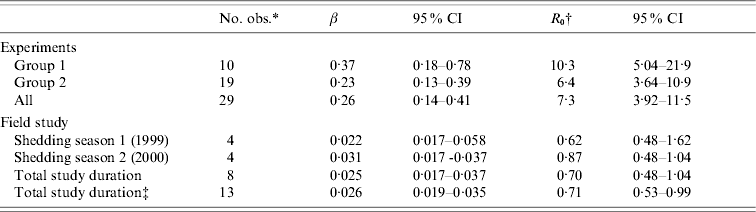
* Number of observations (time intervals) that could be used in GLM model.
† Calculated using an assumed shedding duration of 28 days.
‡ Assumption: certain extent of infectivity remains during the non-shedding season; I+1, S – 1 for each time interval.
In the field study, R 0 was 0·62 (95% CI 0·48–1·62) for the first shedding season (1999), 0·87 (95% CI 0·48–1·04) for the second shedding season (2000) and 0·70 (95% CI 0·48–1·04) for the total study period (Table 3). Assuming that a certain amount of infectivity remained during the non-shedding season [winter; by considering one animal infectious instead of susceptible (I+1; S – 1) at each sampling interval], the overall estimate remained about the same, i.e. 0·71 (95% CI 0·53–0·99).
DISCUSSION
The aim of this paper was to quantify transmission of O157 VTEC based on results of a longitudinally sampled dairy cattle farm known to be positive for O157 VTEC and on an experiment with calves. Additionally, we investigated the role of previously positive animals and pastures in initiating infection in the next shedding season.
The estimate of R 0 from the transmission experiment with calves, was 7·3 (significantly greater than 1), indicating that each infectious calf can infect seven other calves on average, during an assumed infectious period of 28 days in a fully susceptible population; thus probably leading to a major outbreak. In contrast, R 0 in a dairy herd was 0·70 (not significantly less than 1). Because some assumptions underlying transmission models could not be validated in our study, these estimates should be considered an approximate quantification of transmission in cattle. Laegreid & Keen [Reference Laegreid and Keen24] estimated R 0 for O157 VTEC to be 5·25 (95% CI 3·87–6·64) in beef calves using the final size of the infected population, based on serology. Previous research [Reference Laegreid, Elder and Keen31] indicated that the proportion of animals shedding O157 VTEC in faeces was substantially lower (7·4%; min. 0%, max. 20%) than the proportion of animals showing a positive antibody response (83·7%; min. 63%, max. 100%). It is not clear whether calves in which O157 VTEC passes through the gastrointestinal tract, but do not necessarily become infectious shedders, might also show seroconversion. In that case, these calves could be misclassified, leading to overestimation of R 0 when antibody titres are used to estimate transmission rates.
An important question for interpreting results is when to consider an animal to be infectious. In the transmission experiment, we observed continuous shedders and intermittent shedders. For the transmission experiment, therefore, we assumed that a calf was infectious when it tested positive at any sampling, followed by two consecutive positive samplings. Similarly, we assumed that a calf was susceptible when it is tested negative at any sampling, followed by two consecutive negative samplings). However, in an alternative analysis we considered a calf to be infectious when it tested positive and to be susceptible when it is tested negative and almost no difference was found in the R 0 values. We did not quantify the concentration of O157 in faeces, which might affect the transmission dynamics assuming high shedders to be more infectious [Reference Matthews26]. It is possible that continuous shedders in this experiment were also shedding larger numbers of O157 VTEC.
Because there is seasonal variation in cattle shedding O157 VTEC [Reference Heuvelink13–Reference Garber19], transmission rates might differ by season. Modelling a seasonal effect in calculating transmission rates, might better mimic reality. However, we had insufficient data to do this. The estimate of β might be considered as a transmission rate for summer/early autumn (the period in which the experiment was carried out) in calves and might differ substantially from the estimate of β for winter. For the dairy herd, therefore, we also modelled transmission assuming a fixed number of infectious animals, and assumed them to be undetected, present in the population during winter, implying that the infection continued during winter. By adding 1 to numbers for I (I+1) and subtracting 1 from numbers for S (S – 1) for each time interval, five additional intervals could be used in the analysis, thus increasing the power of the analysis. However, estimates of this analysis were similar.
Because the length of the infectious period was unknown and because the excretion pattern varied widely among animals [Reference Brown20–Reference Zhao23, Reference Sanderson32], it was difficult to determine when the infection chain had ended. To calculate R 0 in this study and to make a conservative comparison between calves and adult cattle, we assumed the infectious period for a calf or cow was 28 days on average. The sampling intervals in the longitudinal field study were long compared with the assumed length of the infectious period. This means that some short-lived infections could have been missed in the analysis within a specific sampling interval. However, we might have missed not only cases (C), but also the resulting infectious (I) individuals, resulting in a similar mistake in both the numerator and denominator of R 0. Theoretical reasoning would suggest, therefore, that that there would not be a difference in point estimate of R 0, but only in the confidence interval.
Individuals were reported to shed up to 100 days for adult cows and even up to 189 days for calves [Reference Stewart and Flint33]. After the transmission experiment, when shedding calves were housed individually, O157 VTEC was detected for about 40 days. The mean infectious period for both cows and calves might therefore exceed 28 days. For cows, when the average infectious period exceeds 38 days, R 0 would become >1, enabling major outbreaks to occur. For calves, however, R 0 is already >1, with an infectious period of only 5 days. This indicates that more accurate information is needed about the length of the infectious period, especially for dairy cows, to accurately estimate R 0. Furthermore, because calves are known to have much larger infectious periods than cows, and based on estimates of transmission coefficients from this study, a large difference between R 0 of calves and cows is likely to exist.
Estimated infection rates for the transmission experiment were about ten times greater (R 0=7·3) than those for the field study (R 0=0·70, which might have several causes. Calves, experimentally inoculated with rather large doses, were expected to excrete more than the naturally colonized cows in the field study. The different estimates of transmission coefficients (β) suggest the need for a closer investigation of the role of dose in E. coli O157 infections. Even when weaned calves and cows were infected with identical doses, calves shed larger numbers of O157 VTEC for a longer period than the older animals [Reference Cray and Moon21], possibly indicating an effect of age. In the field study, only data of cows were used as input in the statistical model, because young stock only were sampled during winter and this probably resulted in an overall lower transmission rate because young stock represent better shedders.
In addition, experimental design of the transmission experiment may also have had an effect. The circumstances in the experiment were more controlled than those in the field study, e.g. resulting in a smaller role for the environment. Our results in the field study confirmed that O157 VTEC were able to survive in the environment of cattle and in other animals [Reference Schouten16]. In the field study, a combination of several factors (animals and environment, e.g. cattle and pastures) might play a role in the dynamics of O157 VTEC. In our present experimental study, only cattle data were included in the statistical transmission model, because the infectivity of pastures (especially over winter) was not confirmed. Furthermore, when starting sampling of the herd (summer), the O157 VTEC outbreak appeared to be at its peak and declining. As a result, a low R 0 was calculated for the first shedding season, although the outbreak in the first season seemed to have affected relatively more animals than the outbreak in the second. When starting sampling at the beginning of the shedding season, possibly more contact infections would have been detected. Calves in the experiment were fed supplemental concentrates in a feeding trough on the ground, where calves frequently placed their claws while eating. Faecal contamination of feed could easily have occurred, leading to (indirect) transmission. Cows were fed roughage from a feeding bunk and concentrates from a feeding dispenser, both of which were less easily contaminated with faeces.
Some calves that tested positive in the transmission experiment seemed to start shedding again after a period of testing negative, and some cattle in the field study also tested positive more than once during a shedding season. For this reason, we assumed no transfer of immunity from previous colonization when calculating the reproduction ratios. Cattle that were infected and shedding in the first season, however, did not test positive for O157 VTEC in the second season [Reference Schouten16], so shedding of O157 VTEC seemed to have limited itself to one shedding season. Although this limitation to one shedding season might be coincidental it may be that a certain level of immunity was acquired, induced by the infection in the first season. Potter et al. [Reference Potter34] established that cattle were able to develop immunity for some virulence factors of E. coli O157:H7, i.e. secreted proteins that are assumed to play a role in colonization of host epithelial cells. In the long term, therefore, some resistance against O157 VTEC might have developed, which might have influenced estimates of R 0. Calves infected in the transmission experiment and put out in a ‘clean’ pasture a year later, did not start shedding again. It seems, therefore, that these calves did not function as a reservoir of E. coli O157. Because soil samples tested negative for E. coli O157, calves were probably not exposed to E. coli O157. Therefore, no conclusion can be made, about long-term resistance against the inoculated type of O157 VTEC the previous year before. Whether or not these calves would have started shedding again after experimental challenge with the same or a different strain remains unknown.
After susceptible calves grazed on a pasture infected with O157 VTEC the previous year, no infection occurred. Allowing a pasture to lie fallow for the winter therefore seems to be sufficient to prevent spread to susceptible animals the next spring. For economic reasons, calves used for this experiment were bulls. Research has shown no effect of sex on faecal prevalence and shedding of O157 VTEC [Reference Looper35–Reference Donkersgoed, Graham and Gannon37].
In summary, previously infected calves and contaminated pastures did not contribute to possible infection in the next shedding season. Our results indicate that transmission of O157 VTEC occurs both in experimentally and naturally infected cattle. Transmission rates differ tenfold between weaned calves and dairy cows. Control strategies to reduce the infection rate probably have more impact in calves than in cows. Therefore, to reduce the number of infected animals on a farm one should look for on-farm measures that reduce transmission within calves or young stock. This recommendation is in agreement with that of Turner et al. [Reference Turner25] on the basis of a differential equation model that described transmission in a multigroup managed herd. In their model, however, the dynamics of the infectious organism itself was described rather than the dynamics of infectious and susceptible animals.
ACKNOWLEDGEMENTS
This study was a joint project of Wageningen University and the National Institute of Public Health and the Environment (RIVM). The authors thank the employees of ID–Lelystad, and M. van Eckeveld for their assistance in the collection of faecal and blood samples. Thanks are also due to W. Vullings, E. Koeleman and J. de Bree for their assistance. The support and technical assistance from A. van Zijderveld is greatly appreciated.
DECLARATION OF INTEREST
None.






