Introduction
Spodoptera litura (Lepidoptera: Noctuidae) is an important agricultural pest feeding on host plants covering more than 120 species, including many commercial crops, cotton, soybean, tobacco, cruciferous vegetable, and so on (https://www.cabidigitallibrary.org/doi/10.1079/cabicompendium.44520). S. litura is also a gluttonous insect with high fecundity and mobility, and easy to break out. As the frequent chemical control, S. litura has developed resistance to 42 chemical insecticides (https://www.pesticideresistance.org/display.php?page=species&arId=282), especially to pyrethroids, organophosphates and carbamates (Ahmad et al., Reference Ahmad, Sayyed, Saleem and Ahmad2008; Wan et al., Reference Wan, Wu, Huang, Yu and Wu2008; Saleem et al., Reference Saleem, Hussain, Ghouse, Abbas and Fisher2016; Wang et al., Reference Wang, Huang, Hao, Ran, Wu, Cui, Yang, Jiang and Yang2018; Xu et al., Reference Xu, Mei, Liu, Chen, Li and Wang2020; Zhang et al., Reference Zhang, Gao, Qu, Gong, Li, Luo and Wang2022). Pyrethroids showed strong insecticidal activity, low toxicity to mammals, broad spectrum and excellent stability. Cyhalothrin, β-cypermethrin and fenvalerate are conventional pyrethroids widely used in controlling pests in agriculture, public health and graziery. Metabolism resistance mediated by the detoxification enzyme of cytochrome P450 systems (P450s), carboxylesterases (CarEs) and glutathione S-transferases (GSTs) are proved to be an important reason leading to the resistance to pyrethroids, organophosphates, and carbamates (Li et al., Reference Li, Schuler and Berenbaum2007).
CarEs genes in insects are divided into 14 clades (A-N) showing functions in three aspects, dietary detoxification, pheromone/hormone degradation and neurodevelopment (Oakeshott et al., Reference Oakeshott, Claudianos, Campbell, Newcomb, Russell, Gilbert, Iatrou and Gill2005). The main CarEs clades included α-esterases, juvenile hormone esterases, β-esterases, integument esterase, gliotactins, acetylcholinesterases, neurotactins, neuroligins, glutactin, and uncharacterised group (Oakeshott et al., Reference Oakeshott, Claudianos, Campbell, Newcomb, Russell, Gilbert, Iatrou and Gill2005). CarEs genes in α-esterase, juvenile hormone esterase and β-esterase families accounted for the majority of catalytically active CarEs (Ranson et al., Reference Ranson, Claudianos, Ortelli, Abgrall, Hemingway, Sharakhova, Unger, Collins and Feyereisen2002). CarEs genes in neurotactin, neuroligin, gliotactin, and glutactin are considered to be noncatalytic, and play essential roles in cell-to-cell interactions (Oakeshott et al., Reference Oakeshott, Claudianos, Campbell, Newcomb, Russell, Gilbert, Iatrou and Gill2005), pheromone degradation in moths (Vogt, Reference Vogt, Gilbert, Iatrou and Gill2005) and hydrolysis of the neurotransmitter acetylcholine and juvenile hormone (Taylor and Radic, Reference Taylor and Radic1994; Riddiford et al., Reference Riddiford, Hiruma, Zhou and Nelson2003). As a detoxification enzyme in insects, CarEs function as phase Ⅰ enzyme and could catalytic hydrolysis insecticides contain carboxyl ester by gene amplification, upregulation and coding sequence mutations (Bass et al., Reference Bass, Puinean, Zimmer, Denholm, Field, Foster, Gutbrod, Nauen, Slater and Williamson2014; Wang et al., Reference Wang, Huang, Lu, Jiang, Smagghe, Feng, Yuan, Wei and Wang2015; Hopkins et al., Reference Hopkins, Fraser, Mabbitt, Carr, Oakeshott and Jackson2017). The CarEs involved hydrolyse reaction is divided into two steps. Firstly, a nucleophilic attack by the oxygen of serine residue at the active site is taken on the carbonyl carbon of the substrate, releasing an alcohol product and forming an acyl-enzyme. Secondly, another nucleophilic attack is taken by water, releasing an acid product and forming the free enzyme (Oakeshott et al., Reference Oakeshott, Claudianos, Campbell, Newcomb, Russell, Gilbert, Iatrou and Gill2005).
In S. litura, multiple studies have implied the involvement of CarEs in insecticide resistance by synergist experiments (Armes et al., Reference Armes, Wightman, Jadhav and Rao1997; Ahmad et al., Reference Ahmad, Arif and Ahmad2007; Huang and Han, Reference Huang and Han2007), increased enzyme activities (Li et al., Reference Li, Zhu, Shan, Li, Liang and Gao2021a), or overexpression of CarEs genes in resistant strains (Xu et al., Reference Xu, Mei, Liu, Chen, Li and Wang2020). Only a few researches provided the evidence proving the involvement of CarEs genes in insecticide resistance (Shi et al., Reference Shi, Li, Zhou, Liao and Shi2022; Li et al., Reference Li, Lv, Liu, Bi, Pan and Shang2023). The function of each specific CarEs genes in insecticides resistance is still lacking.
In our previous study, the addition of CarEs synergist, triphenyl phosphate (TPP), significantly increased the mortality of larvae from a field-collected population after pyrethroids and organophosphates treatment, suggesting a possible role of CarEs genes in pyrethroids and organophosphates resistance (Li et al., Reference Li, He, Xie, Kong, Wu, Shi, Liu and Xu2021b). A series CarEs genes were found to show significantly higher reads in the field-collected population resistant to pyrethroids and organophosphates than the susceptible population by RNA-Seq (data derived from NCBI Sequence Read Archive database with accession number PRJNA843172, Table S1, Xu et al., Reference Xu, Li, Liu, Zhang, Liu, Yu and Li2022). SlCarE054 was selected to heterologous expressed due to its high expression level in the resistant population. Its metabolism activity to pyrethroids and organophosphates was determined by ultra performance liquid chromatography (UPLC), and the metabolite was identified by Gas Chromatography-Mass Spectrometer (GC-MS). The interaction between insecticides and SlCarE054 was also conducted by molecular docking. Our results enriched the knowledge of the role of CarEs genes in insecticides resistance, and explained the CarEs involved molecular mechanism to pyrethroids and organophosphates resistance in S. litura.
Materials and methods
Insects and chemicals
The susceptible population (GX) was firstly collected from Nanning, Guangxi province, China and provided by Guangxi Tianyuan Biochemistry Corp., Ltd. QJ population, showing extremely high-level resistance to fenvalerate (Resistance ratio, RR>25,000), β-cypermethrin (RR = 3659- fold), cyhalothrin (RR>50,000), phoxim (RR = 295- fold) and chlorpyrifos (RR = 207- fold) compared with GX population (Li et al., Reference Li, He, Xie, Kong, Wu, Shi, Liu and Xu2021b), was firstly collected from Qianjiang, Hubei province, China at 2019. KX population was collected from Xinxiang, Henan province, China at 2022. The rear condition was: 26 ± 1°C, 60–70% relative humidity, 14:10 h of light:dark photoperiod.
The technical grade of fenvalerate (93.4%) was provided by Jiangsu Changlong Chemicals Co., Ltd. The technical grade of β-cypermethrin (95.0%), cyhalothrin (98.4%), phoxim (91.0%) and chlorpyrifos (95.0%), were obtained from Beijing Huarong Biochemical Co., Ltd.
Bioassay was determined by topical application described in Xu et al. (Reference Xu, Mei, Liu, Chen, Li and Wang2020). Briefly, a series diluted insecticides were prepared in acetone and one microliter of insecticides was applied on the thoracic dorsum of third instar larvae of S. litura from GX and KX populations. The mortality of larvae was checked 48 h after treatment. Application of acetone was used as control. Each concentration consisted of three replicates.
Cloning the coding sequence of SlCarE054 and its bioinformatics analysis
The total RNA from third instar larvae was extracted by TaKaRa MiniBEST Universal RNA Extraction Kit (Takara, Dalian, China). cDNA was synthesised using the FastQuant RT Kit (with gDNase) (Tiangen, Beijing, China). The coding sequence of SlCarE054 was cloned with primer pairs in Table S2. The PCR product was purified by TIANgel Midi Purification Kit (Tiangen) and inserted to pLB vector (Tiangen), then transformed into E. coli DH5α competent cells. Positive clones were selected and sequenced at Sangon Biotech (Shanghai, China).
The protein molecular weight and theoretical isoelectric point (pI) of SlCarE054 were predicted by Compute pI/Mw tool (https://web.expasy.org/compute_pi/). The signal peptide and protein glycosylation were detected by SignalP-6.0 (https://services.healthtech.dtu.dk/services/SignalP-6.0/) and NetNGlyc1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/), respectively. The amino acid sequence of SlCarE054 was blasted with CarEs genes in other lepidoptera insects using GENEDOC software (https://www.nrbsc.org/gfx/genedoc/). The conserved motifs of SlCarE054 were analysed by CD-search in NCBI (https://www.ncbi.nlm.nih.gov/cdd/?term=).
Determination the overexpression of CarEs genes and spatiotemporal expression of SlCarE054
The expression of CarEs genes in QJ, KX, and GX population was validated by quantitative RT-PCR (qRT-PCR). RNA samples from the third instar larvae of QJ, KX, and GX populations were extracted and the relative expression level of SlCarE054 was quantified using SuperReal PreMix Plus (SYBR Green, Tiangen) on ABI QuantStudio 6 PCR System (Applied Biosystems by Life Technologies, Foster, CA, U.S.A.).
To determine the spatiotemporal expression of SlCarE054, RNA samples from the first, second, third, fourth, fifth, sixth instar larvae and pupae, tissues including head, cuticle, foregut, midgut, hindgut, malpighian tubules, fat body from the fifth instar larvae of QJ population were extracted and also quantified by qRT-PCR. The reaction system consisted of 2 × SuperReal PreMix solution (10 μl), 50 × ROX reference dye (0.4 μl), each of an F and R primer (10 μmol l−1, 0.6 μl, Table S2), cDNA (1 μl), and ddH2O (7.4 μl). The thermal cycling was conducted according to the protocols. EF-1α and RPL10 were used as housekeeping genes (Lu et al., Reference Lu, Yuan, Gao, Kang, Zhan, Wan and Li2013). The reaction of each gene was repeated with 3 independent mRNA samples. The relative expression of SlCarE054 in each sample was quantified by 2−ΔΔCt method.
The heterologous expression of SlCarE054 in E. coli
The coding sequence of SlCarE054 (deleting the signal peptide region) was digested by BamH I and Xho I (Takara, Dalian, China), and then subcloned into the expression vector pET-30a(+). The recombinant plasmid pET-30a(+)/SlCarE054 was transformed into E. coli BL21 (DE3) and induced by isopropyl β-D-thiogalactoside (IPTG, 1.0 mmol l−1) at 18°C, 160 rpm for 48 h. The mixture was centrifuged at 4°C, 6000 rpm for 10 min and the cells were transferred on ice to lyse by ultrasonic (Sonics and Materials, Inc., U.S.A.). The crude enzyme was obtained from the supernatant after centrifuging at 4°C, 16,000 rpm for 30 min and further purified by ProteinIso® Ni-NTA Resin (TransGen Biotech, Beijing, China) with 100 and 200 mmol l−1 imidazole, desalted by Zeba™ Spin Desalting Columns (Thermo, Shanghai, China).
The extracts of empty vector and recombinant protein SlCarE054 were checked by SDS-PAGE. The enzymatic activity of recombinant protein SlCarE054 was detected using α-naphthyl acetate (α-NA) as the model substrate. The stock solution of α-NA was dissolved in acetone. A series concentration of α-NA working solution (12.5, 25, 50, 100, 200 μM) was diluted by Fast Blue RR salt. The purified recombinant enzyme (33 μl, 0.5 mg ml−1) was added with PBS (297 μl), α-NA (330 μL) and mixed well immediately. The mixture was transferred to 96-well microplates, and the absorption at 450 nm was determined every 1 min at 30°C for 5 min in a time-driver model by microplate reader (Biotek, U.S.A.). Km and Vmax values of recombinant protein SlCarE054 were determined according to the Michaelis–Menten equation with SigmaPlot 12.0 (Systat Software, San Jose, CA, U.S.A.).
Insecticides metabolism by recombinant protein SlCarE054 and the identification of metabolites
Purified recombinant protein SlCarE054 (50 μl, 0.5 mg ml−1) was added with the mixture containing PBS (400 μL, 0.1 mol l−1, pH 7.0) and insecticides (450 μl, 50 mg l−1) diluted with PBS. Boiled purified recombinant protein SlCarE054 was used as control. The reaction was incubated at 30°C for 2 h and stopped by adding acetonitrile (900 μl). The residual insecticides were extracted by adding NaCl to saturation, shaking at 160 rpm for 2 h, vortexing for 5 min, and centrifuging at 12,000 rpm for 8 min. The supernatant was filtered by 0.22 μm membrane and quantified by UPLC (Waters Acquity UPLC system, U.S.A.) using the detection method in Li et al. (Reference Li, He, Xie, Kong, Wu, Shi, Liu and Xu2021b). Briefly, acetonitrile/water (80:20, v/v) was used as mobile phase with flow rate at 0.4 ml min−1. The injection volume was 3 μl. The residual quantity of fenvalerate, β-cypermethrin, cyhalothrin, phoxim, and chlorpyrifos was detected using a PDA detector at 220, 220, 230, 280, and 290 nm, respectively.
The metabolites were identified using GC/MS (Agilent 7890B/5977A, Santa Clara, CA, U.S.A.) equipped with a capillary column (HP-5MS, 0.25 μm × 250 μm × 30 m) and mass database of NIST14.L. The injection volume was 1 μL in splitless mode using carrier gas of helium (≥99.999%) with flow rate at 1 ml min−1. The temperature of inlet was 290°C. The initial oven temperature was set at 60°C for 1 min, with a subsequent temperature gradient of 20°C per minute until a final temperature of 290°C. The transfer line temperature was 290°C. The conditions of mass spectrometry were as follows: the ion source was in the EI mode, 70 eV, 230°C. The temperature of quadrupole was 150°C, and the mass scanning range was 30–500 MHZ.
Homology modelling and molecular docking
The 3D protein structure of SlCarE054 was constructed online using Swiss model server (https://swissmodel.expasy.org/, Bienert et al., Reference Bienert, Waterhouse, de Beer, Tauriello, Studer, Bordoli and Schwede2017; Waterhouse et al., Reference Waterhouse, Bertoni, Bienert, Studer, Tauriello, Gumienny, Heer, de Beer, Rempfer, Bordoli, Lepore and Schwede2018). The carboxylic ester hydrolase from S. frugiperda (PDB ID: A0A2H1V6U9.1.A) with 84.95% sequence identity was selected as template. The 3D structure of fenvalerate, β-cypermethrin, cyhalothrin, phoxim, and chlorpyrifos were sketched and obtained by ChemDraw 20.0 (PerkinElmer, U.S.A.) and Chem3D (PerkinElmer, U.S.A.). Autodock 4.2 (Autodock, San Diego, CA, U.S.A., Morris et al., Reference Morris, Huey, Lindstrom, Sanner, Belew, Goodsell and Olson2009) was used to perform the molecular docking with the default parameters. The possible interactions and the binding free energy between ligand and protein were predicted by Autodock 4.2. The interaction between ligand and protein was further predicted using PLIP (Protein-Ligand Interaction Profiler, https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index). The results of modelling and docking were visualised by PyMOL (New York, NY, U.S.A.).
Statistical analysis
The expression of CarEs genes in QJ, KX, and GX populations, and the in vitro metabolism activity of SlCarE054 were evaluated by Students' t-test. The relative expression of SlCarE054 in different development stage and tissues were analysed by One-way analysis of variation (ANOVA) followed by Tukey's HSD multiple comparisons test. P < 0.05 was regarded to be statistically significant.
Results
Kx population showed high resistance level to pyrethroids and organophosphates
As shown in table 1, the LD50 values of KX population to fenvalerate, cyhalothrin, β-cypermethrin, chlorpyrifos, and phoxim were 108.200, 73.000, 3.295, 10.340, and 3.090 μg larva−1, respectively. Compared with GX population, the resistance ratios of KX population to fenvalerate, cyhalothrin, β-cypermethrin, chlorpyrifos, and phoxim were 15,457.14-, 73,000.00-, 1657.50-, 229.78-, and 99.68- fold, respectively.
Table 1. The resistance ratio of S. litura from KX and GX populations to pyrethroids and organophosphates.
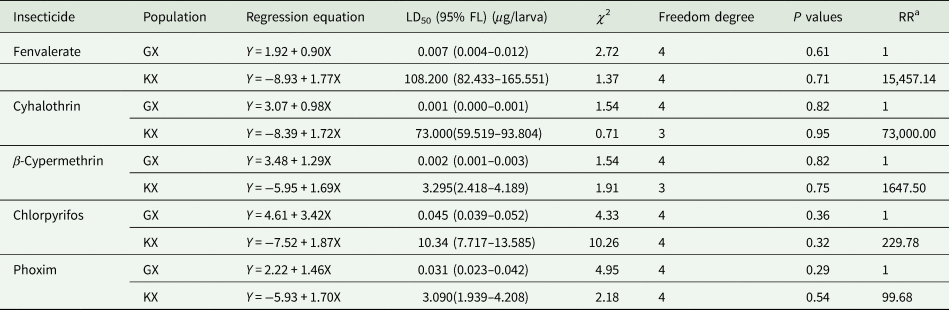
a RR = LD50 value of KX population/LD50 value of GX population.
The sequence information of SlCarE054 and its bioinformatics
The cDNA full length of SlCarE054 was 1677 bp, encoding 558 amino acids (Table S3). The protein molecular weight and theoretical pI of SlCarE054 were predicted to be 61.04 kDa and 5.45, respectively. According to the SignalP-6.0, the first 18 amino acids at the N-termini of SlCarE054 were signal peptides (fig. S1). There are three predicted N-glycosylation sites: Asn116-Leu117-Ser118-Val119, Asn256-Phe257-Thr258-Asp259 and Asn459-Leu460-Thr461-His462. The conserved domain analysis identified the catalytic triad (Ser205, Glu334, and His447), the substrate binding pocket (Gly125, Gly126, Gly127, Tyr204, Ser205, Ala206, Ala209, Phe356, Asp360, Leu361, Phe398, Cys448, Ile452), and the pentapeptide motifs (Gly203-X-Ser205-X-Gly206, fig. 1).

Figure 1. The alignment of SlCarE054 with Ha001A (AMO44416.1), Ha001 G (AMO44418.1), Ha001H (AMO44417.1) in Helicoverpa armigera, PxαE8 (XP_048485366.1), PxCCE016b (AIN76405.1) in Plutella xylosterlla, and SlCarE074 (XP_022837633.1) in S. litura. The signal peptide sequences of SlCarE054 were indicated in underline with blue line, the catalytic triad residues were marked with triangles, the highly conserved pentapeptide residues are marked with green horizontal rectangle, the potential N-glycosylation was underlined with green lines, the substrate binding sites are labelled with red underlines.
Validation of the overexpression of CarEs genes and the spatiotemporal expression of SlCarE054 in QJ population
A total of 11 significantly up-regulated CarEs genes in QJ population were selected from the RNA-Seq result. The expression of five CarEs genes (SlCarE054, SlCarE056, SlCarE004, SlCarE020, SlCarE077) were both overexpressed in QJ and KX populations validated by qRT-PCR (fig. 2). SlCarE054 showed the highest relative expression level in QJ population (7.60 ± 1.25) and was thus selected for further study.
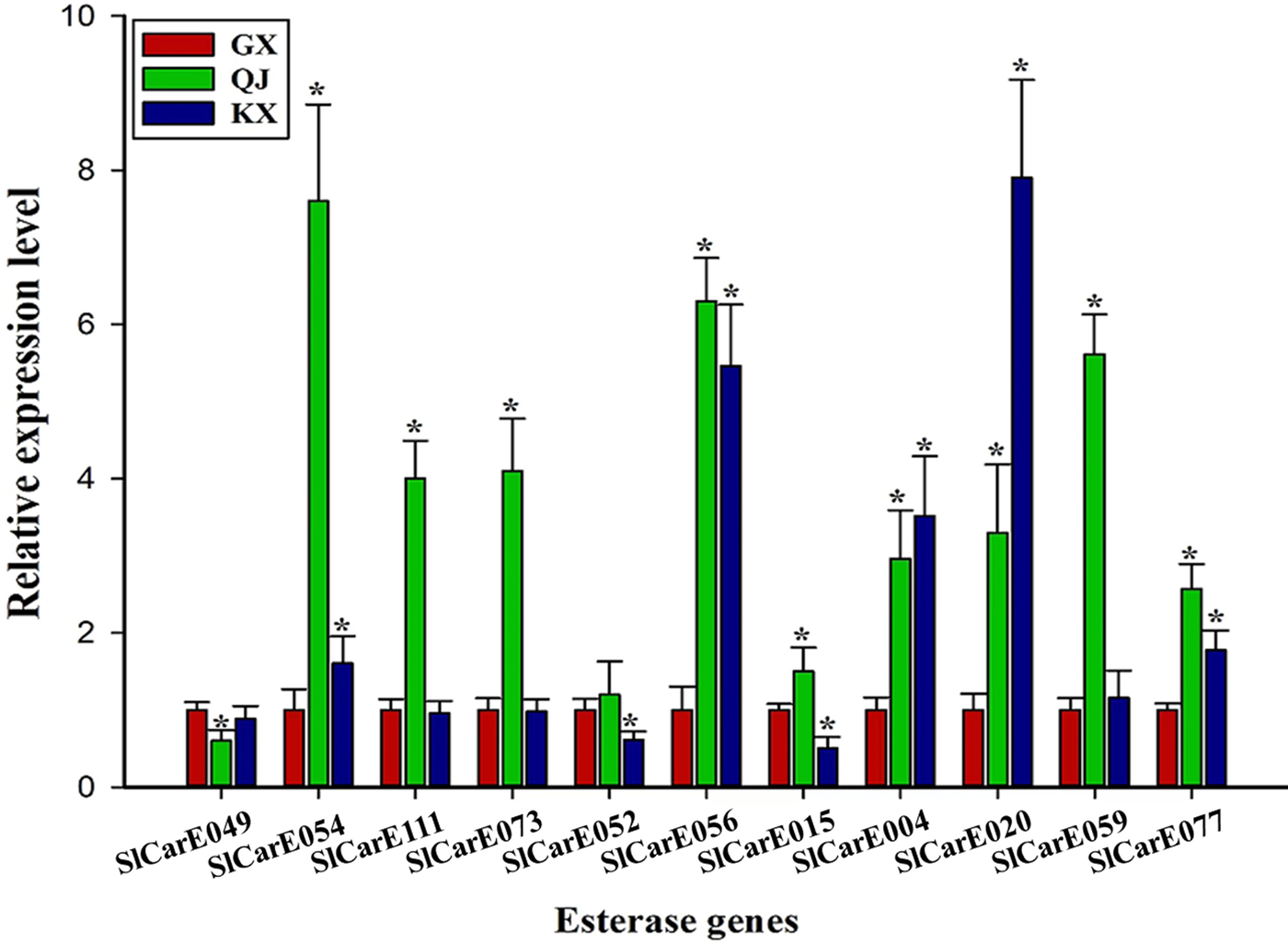
Figure 2. The relative expression level of CarEs genes in QJ, KX, and GX populations validated by qRT-PCR. ‘*’ indicates significant difference compared with GX population analysed by Students' t-test (P < 0.05).
The relative expression of SlCarE054 in the larvae and pupae stage of QJ population was determined. The expression of SlCarE054 showed no significant change during the larvae stage, while increased significantly in the pupae, which were 2.8- fold than that in the first instar larvae (fig. 3A). Among the tissues in the fifth instar larvae, SlCarE054 showed the lowest expression in the head, cuticle, and fat body. Its expression in the midgut and foregut were significantly higher than that in head, which were 46.3- and 21.3-fold, respectively (fig. 3B). The expression of SlCarE054 in the hindgut and malpighian tubules were significantly higher than that in head or fat body, and showed no significant difference with that in cuticle.

Figure 3. The spatiotemporal expression of SlCarE054 in QJ population. (A) The expression of SlCarE054 in the developmental stages of larvae. (B) The relative expression level of SlCarE054 in different tissues of the fifth instar larvae. Different lowercase letters above the bars indicated significant difference analysed by ANOVA followed by Tukey's HSD test (P < 0.05). Developmental stages: the first instar larvae (L1), the second instar larvae (L2), the third instar larvae (L3), the fourth instar larvae (L4), the fifth instar larvae (L5), the sixth instar larvae (L6), pupae (PU). Tissues: head (H), cuticle (C), foregut (FG), midgut (MG), hindgut (HG), malpighian tubules (MT), fat body (FB).
Identification of recombinant protein SlCarE054 and its enzymatic activity to α-NA
The extracts of recombinant protein were identified by SDS-PAGE. The bands of extracts induced by IPTG and purified recombinant protein were shown nearly to 66.2 kDa in the lanes (fig. 4A). This result was well coincided with the predicted molecular weight of 61.04 kDa. The quantity of recombinant protein purified by imidazole at 100 mmol l−1 is more abundant than that of 200 mmol l−1. Thus, imidazole at 100 mmol l−1 was used for recombinant protein purification.

Figure 4. Characterisation of the recombinant protein SlCarE054 by SDS-PAGE (A) and the kinetic parameters of recombinant protein SlCarE054 (B). Marker: protein marker; pET30a(+), extract of vector only; IPTG-, extract of the recombinant protein without IPTG induction; IPTG + , extract of the recombinant protein induced by IPTG; 100 mM, the purified recombinant protein induced by IPTG eluted by 100 mM imidazole; 200 mM, the purified recombinant protein induced by IPTG eluted by 200 mM imidazole. Km and Vmax values of SlCarE054 were calculated using α-NA as substrate with Michaelis–Menten plot.
The esterase activity of purified recombinant protein SlCarE054 to α-NA was determined. The Vmax and Km values of SlCarE054 were calculated to be 38.1 ± 6.1 OD min−1 mg−1 and 59.5 ± 8.3 μM, respectively (fig. 4B).
The metabolic activity of SlCarE054 to pyrethroids and organophosphates
The metabolic activity of SlCarE054 to pyrethroids and organophosphates was determined using the purified recombinant protein by UPLC following incubation with insecticides for 2 h. Results showed that the insecticides residue area of the determined pyrethroids incubated with purified recombinant protein SlCarE054 decreased significantly than that incubated with boiled SlCarE054. As a chiral insecticide, β-cypermethrin was resolved into α-cypermethrin and θ-cypermethrin under the detection condition. The depletion rate of SlCarE054 to θ-cypermethrin was 33.8%, the highest among all the pyrethroids, while 7.3% to α-cypermethrin, obviously lower than the enantiomer θ-cypermethrin. The depletion rate of SlCarE054 to cyhalothrin and fenvalerate were 6.4 and 5.2%, respectively (fig. 5). After incubated with SlCarE054, the insecticides residue area of phoxim or chlorpyrifos remain unchanged.

Figure 5. The metabolic activity of recombinant protein SlCarE054 to organophosphates and pyrethroids. ‘*’ indicates significant difference analysed by Students' t-test (P < 0.05). The number above bars indicated the depletion rate of insecticides metabolised by SlCarE054.
The metabolites of β-cypermethrin after incubation with SlCarE054 were determined by GC/MS. According to the peak, the possible metabolites and degradation pathways were deduced. The ester bond of β-cypermethrin was hydrolysed and the corresponding alcohol was generated. The alcohol was further metabolised to 3-phenoxybenzaldehyde (fig. S7).
Predication on the interaction of SlCarE054 with pyrethroids and organophosphates by molecular docking
The 3D structure of SlCarE054 included 14 α-helixes, 13 β-sheets and connected by several loops. An obvious loop at the N-terminal indicated the signal peptide (fig. 6). β-cypermethrin could bind to the residues of catalytic triad Ser205 and His447 of SlCarE054, and form hydrogen bonds with them. Another hydrogen bond was predicted to bind with residue Gly126 with 3.4 Å. Hydrophobic interactions were predicted to bind with the residues Leu460, Tyr204, Phe285, Tyr341, Met337, His447 with distance 3.0–4.5 Å. Molecular docking results showed that cyhalothrin and fenvalerate bind to the sites near to the catalytic triad of SlCarE054. The interaction to fenvalerate included a hydrogen bond with residue Leu460, a π-stacking with residue His447, and hydrophobic interactions with residues Thr458, Phe472, Asp449, Met337, His447, Ile452 with distance 3.2–3.7 Å. The interaction with cyhalothrin included a hydrogen bond with residue Asn446, and hydrophobic interactions with residues Thr434, Pro435, Phe472, Asp449, Leu336, Leu339 with distance 3.3–4.0 Å. These residues were mostly near to the catalytic triad, His447 and Glu334.

Figure 6. The interactions of SlCarE054 with pyrethroids and organophosphates. β-cypermethrin, cyhalothrin, fenvalerate, chlorpyrifos, and phoxim were indicated by yellow, green, fuchsia, blue, and peridot, respectively. The blue lines indicate the hydrogen bond; the gray lines indicated the hydrophobic interaction; the green lines indicate the π-stacking.
The residues of SlCarE054 interacted with phoxim or chlorpyrifos were clearly different with those of pyrethroids. The interaction with chlorpyrifos included a hydrogen bond with Met135 and a π-stacking with Trp132. For phoxim, there are two hydrogen bonds with Trp132 and Leu460, a π-stacking with Trp132, hydrophobic interactions with Asp456, Leu460, and Pro463. These residues were far away from the catalytic triad.
The free binding energy of SlCarE054 to β-cypermethrin, fenvalerate, cyhalothrin, phoxim, and chlorpyrifos were predicted to be −7.19, −6.94, −6.78, −6.27, and −5.18 Kcal mol−1, respectively.
Discussion
In the genome of S. litura, a total of 110 CarEs genes have been identified. CarEs genes from lepidoptera and α classes showed large expansions. Most CarEs genes from lepidoptera and α classes could be induced by xanthotoxin and imidacloprid. Silencing of two CarEs genes from lepidoptera class by siRNA injection, larvae treated by imidacloprid showed increased mortality compared with control, indicating CarEs genes expansion contributed to the increased detoxification (Cheng et al., Reference Cheng, Wu, Wu, Chilukuri, Huang, Yamamoto K, Feng, Li, Chen ZW, Liu JQ, Wang XX, Liu DL, Fu BH, Liu C, Tomar A, Montagné N, Alençon E, Bhatnagar RK, Shiotsuki T, Promboon A, Arunkumar KP, Goldsmith MR and Xia QU2017). Meanwhile, five (SlituCOE009, SlituCOE090, SlituCOE050, SlituCOE093, and SlituCOE074) CarEs genes from α-esteras, lepidopteran esterase and integument esterase in the indoxacarb-resistant strains were validated to make contribution to indoxacarb resistance by transcriptional up-regulation (Shi et al., Reference Shi, Li, Zhou, Liao and Shi2022). In this study, five out of eleven selected CarEs genes was found both overexpressed in two field collected S. litura populations (QJ and KX). The relative expression of SlCarE054, SlCarE056, SlCarE077 in QJ and KX populations were agree with their resistance level to determined pyrethroids and organophosphates. SlCarE054, derived from lepidoptera class, showed the highest expression in QJ population among the selected CarEs genes. Its amino acid sequence included a signal peptide with 18 amino acids at the N terminal, which indicated that it was a secreted extracellular protein. The predicted N glycosylation site might enhance protein stability and catalytic efficiency.
SlCarE054 was expressed in all the determined development stages. The expression of SlCarE054 was significantly higher in the digestive tissue midgut. This result was similar with the CarEs genes of Pxαe14 (Li et al., Reference Li, Zhu, Hu, Shi, Qi, Liang and Gao2022), Pxae18, and Pxae28 in Plutella xylosterlla (Xie et al., Reference Xie, Ren, You, Chen, Song and You2017), and CarE001H in H. armigera (Li et al., Reference Li, Bai, Zhao, Xu, Sun, Dong, Li, Liu and Ma2020), which all showed metabolic activity to insecticides, suggesting a possible role of SlCarE054 in insecticide detoxification. While its expression in pupae is significantly higher than that in larvae, which is different from most of the above CarEs genes, similar with that of Pxαe14 (Li et al., Reference Li, Zhu, Hu, Shi, Qi, Liang and Gao2022). This result suggested that SlCarE054 might also play roles in insect physiology during the pupae stage.
CarEs was reported to metabolise pyrethroids and organophosphates by hydrolysing in insects (Tang et al., Reference Tang, Dai, Qi L, Du and Zhang2020). In Musca domestica, the recombinant protein MdaE17, MdaE7, MdbE2, and MdIntE showed significant metabolic activity to permethrin, but no metabolic activity to its metabolite, 3-phenoxybenzylalcohol or 3-phenoxybenzaldehyde, indicating that these CarEs genes only participated in the first step of permethrin degradation (Feng et al., Reference Feng, Li and Liu2018; Feng and Liu, Reference Feng and Liu2020). BoαE1 in Bradysia odoriphaga possesses hydrolase activity towards malathion and participates in the detoxification of malathion (Tang et al., Reference Tang, Dai, Qi L, Du and Zhang2020). CarEs was also reported to participate in organophosphates resistance by sequestering the insecticide away from its target site (tight binding/low turnover, Wheelock et al., Reference Wheelock, Shan and Ottea2005). In Culex Quinquefasciatus, Cqestβ2 confers metabolic resistance by sequestering the organophosphates rather than proceeding via catalytic hydrolysis (Hopkins et al., Reference Hopkins, Fraser, Mabbitt, Carr, Oakeshott and Jackson2017). Lcα-E7 in Lucilia cuprina was reported to sequester the insecticide molecule and then detoxify it slowly, thus mediating a wide range of insecticides resistance (Yan et al., Reference Yan, Cui and Qiao2009; Jackson et al., Reference Jackson, Liu, Carr, Younus, Coppin, Meirelles, Lethierb, Pandeyc, Ollisa, Russellc, Weikb and Oakeshottc2013). The metabolic activity of SlCarE054 to pyrethroids and organophosphates was also determined. Only pyrethroids could be metabolised by SlCarE054, while not the tested organophosphates. SlCarE054 showed the highest metabolic activity to β-cypermethrin, which was similar with the result of CarE001A and CarE001H in H. armigera (Li et al., Reference Li, Bai, Zhao, Xu, Sun, Dong, Li, Liu and Ma2020). The metabolic activity of SlCarE054 to fenvalerate and cyhalothrin were relatively low. This might be explained as that SlCarE054 contribute more to the resistance to β-cypermethrin than that of fenvalerate or cyhalothrin. Also, the metabolic ability of SlCarE054 to β-cypermethrin showed stereoselectivity. The depletion rate of SlCarE054 to θ-cypermethrin is much higher than that of α-cypermethrin. This is similar with the report of stereoselectivity difference of CarEs E4 in Myzus persicae. The purified CarEs E4 could only hydrolyse the 1S-trans-permethrin rapidly, but not the other three isomers (Devonshire and Moores, Reference Devonshire and Moores1982). In M. domestica, docking analysis also found that the isomer of permethrin, 1S-trans-permethrin, fit most snugly within the binding cavities of four CarEs, MdαE7, MdαE17, MdβE2, and MdIntE7 with the lowest binding energy (Feng and Liu, Reference Feng and Liu2020). The metabolite 3-phenoxybenzaldehyde of β-cypermethrin was identified by GC/MS after incubated with SlCarE054 or boiled SlCarE054. The abundant of 3-phenoxybenzaldehyde after incubated with SlCarE054 was much higher than that of boiled SlCarE054, and the abundant of parent compound β-cypermethrin was lower after incubated with SlCarE054 than that of boiled SlCarE054 (fig. S7). Thus, we deduced that SlCarE054 could metabolite β-cypermethrin directly by hydrolysing the ester bond, which was similar with the metabolic pathway of pyrethroids reported in previous studies (Feng et al., Reference Feng, Li and Liu2018; Feng and Liu, Reference Feng and Liu2020). The residual area of phoxim and chlorpyrifos showed no significant change compared with the boiled SlCarE054, suggesting SlCarE054 could not participate in their resistance by direct metabolism or sequestering. As reported by Li et al. (Reference Li, Zhu, Shan, Li, Liang and Gao2021a), the overexpression of PxαE8 in the multi-insecticide resistant P. xylostella populations is involved in the resistance to both β-cypermethrin and phoxim. The overexpression of PxαE14 in P. xylostella participated in the multiple resistance to β-cypermethrin, bifenthrin, chlorpyrifos, fenvalerate, malathion, and phoxim. Also, PxαE14 showed higher metabolic activity to pyrethroids than that of organophosphates, which indicated that PxαE14 may prefer to metabolise carboxyl esters in pyrethroids than phosphate esters in organophosphates (Li et al., Reference Li, Zhu, Hu, Shi, Qi, Liang and Gao2022). This preference might be related with the different structure of proteins.
The molecular docking was further conducted to explain the possible reason for the different metabolic activity to pyrethroids and organophosphates. β-cypermethrin could interact with the residues of conserved catalytic triad (Ser205 and His447) of SlCarE054 and the oxyanion hole (Gly126) by forming hydrogen bonds. Fenvalerate could form π-stacking with residue of catalytic triad His447, and hydrogen bond with Leu460. Cyhalothrin could only form hydrogen bond with Asn446. The binding of pyrethroids with SlCarE054 were also surrounded with several hydrophobic residues around the conserved catalytic triad. These results indicated that pyrethroids could bind to or close to the catalytic triad or substrate binding site, which is beneficial to the occurrence of the first step of the catalytic reaction. The binding free energy of SlCarE054 with pyrethroids follows the order of β-cypermethrin <fenvalerate<cyhalothrin, which could explain the difference in metabolic activity of SlCarE054 to these pyrethroids. The binding free energy of SlCarE054 with organophosphates were much higher than that of pyrethroids, and the interaction sites were away from that of catalytic triad or substrate binding sites.
In conclusion, our results suggested that the overexpression of SlCarE054 in pyrethroids and organophosphates resistant populations could contribute to the resistance to β-cypermethrin by direct metabolism. SlCarE054 showed stereoselectivity to the metabolism of β-cypermethrin.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/S0007485324000282.
Acknowledgements
This work is supported by the Key Scientific and Technological Research Project of Henan Province (222102110044), the Postdoctoral Research Grant in Henan Province (202103118), Young Talent Lifting Project of Henan Province (2023HYTP003), the Henan Provincial Science and Technology Major Project (221100110100), the special fund project for central guiding Henan province local development (Z20221343034).
Competing interest
None.










