Introduction
Twin research methods are genetically sensitive designs which allow researchers to understand the degree to which genetics and environmental factors contribute to variation (individual differences) in variables such as health outcomes. In addition, twin studies allow for associations between traits (disorders) to be quantified in parts due to shared genes and shared environmental factors.
Materials and methods
The Sri Lankan Twin Registry was established in 1997, and is the first ever twin register in South Asia [Reference Sumathipala, Siribaddana, De Silva, Fernando, Abeysingha, Dayaratne, De Silva, Warnasuriya and and Hotopf1]. It is still one of the few large-scale functional population-based twin registries in the developing world [Reference van Dongen, Slagboom, Draisma, Martin and Boomsma2]. In 2007, twins from the registry and a comparative sample of singletons (non-twins) were recruited from Colombo district to conduct a cross-sectional study on mental health titled ‘The Colombo Twin and Singleton Study’ (COTASS 1) [Reference Siribaddana, Ball, Hewage, Glozier, Kovas, Dayaratne, Sumathipala, McGuffin and Hotopf3]. The study was a collaboration between the Institute for Research and Development, Colombo, Sri Lanka (IRD), the Institute of Psychiatry, Psychology and Neuroscience, Kings College London, UK, and Brain and Mind Centre, University of Sydney, Australia. COTASS 1 aimed to bridge a gap in genetic research on mental health in a low- and middle-income country by exploring the genetic and environmental factors involved in common mental disorders in the Sri Lankan population.
The same cohort was revisited in 2012 to conduct a multi-component study on mental health, metabolic syndrome factors, and other health and well-being variables (the Colombo Twin and Singleton follow-up Study – COTASS 2) [Reference Jayaweera, Aschan, Pannala, Adikari, Glozier, Ismail, Pariante, Rijsdijk, Siribaddana, Zavos, Zunszain, Sumathipala and Hotopf4]. The Sri Lankan Twin Registry biobank (SLTR-b) was established as a component of COTASS 2 with the aim to foster innovative epidemiological research. Participant recruitment lasted until the end of December 2015 for all components.
Setting
The COTASS cohort is based in the Colombo district which is a multi-ethnic and multicultural city [5]. Although being the country's capital, it is also the administrative and economic centre of the country. It is the most highly populated district in Sri Lanka with 2.4 million people comprising 11.4% of the country's population. Colombo has a mix of urban and rural areas with the majority being urban. The district is heavily urbanised and more westernised than other areas of Sri Lanka. The population is characterised by great socio-economic diversity in terms of education, employment, and occupational social class.
COTASS 2 comprised of three main study components: surveys, anthropometric measures, and biospecimen collection for clinical investigations and biobanking. This report is an overview of the outcomes, and proceedings of collecting, processing, and storing biospecimen data for the SLTR-b.
Minimising pre-analytical errors
Pre-analytical errors to ensure accurate laboratory results were minimised through careful collection and handling of biospecimens [Reference Hammerling6]. Similar care was taken with biospecimens collected for long-term storage for biobanking such that they would be usable for future downstream processes. A 4-h time frame for biospecimen collection and transportation back to the laboratory was maintained to reduce serum protein concentrations change over time [Reference Flower, Ahuja, Humphries and Mohamed-Ali7, Reference Jackman, Utter, Heitman, Hirschkorn, Law, Gefter, Busch and Norris8]. Pre-printed stickers with participant's unique identification number were used to label collected samples thereby avoiding unreadable or incorrectly written identification numbers. Standard operating procedures for biospecimen collection, storage, and participant preparation instructions for biospecimen collection were developed and optimised during the pilot phase. These minimised variations as biospecimens were collected from multiple sites which included clinical collection centres and house visits.
As infection, inflammatory processes, and medications may influence the outcome of certain blood investigations [Reference Fischbach and Dunning9], participants were excluded from the clinical investigations’ component if they (1) were on long-term medication which could interfere with the parameters under the study (e.g. steroids or other immunomodulatory agents); or (2) suffered from a comorbid inflammatory disease (e.g. chronic infections and autoimmune diseases). Pregnant participants and those suffering from an acute ailment or infection were revisited after delivery or recovery of illness, respectively.
Biospecimen collection
Overnight fasting blood and first morning mid-stream urine was collected contemporaneously from a single participant to measure a variety of clinical investigations and for biobanking (see Table 1 for details). Blood was collected using a vacuum extraction tube system (Becton Dickinson & Co Vacutainer System). Table 1 provides details of the collection order, preservative type, and volumes. Clinical investigations were outsourced to a private hospital in Colombo, housing an accredited laboratory.
Table 1. COTASS 2 collected sample types, collection priority, volume, and purpose
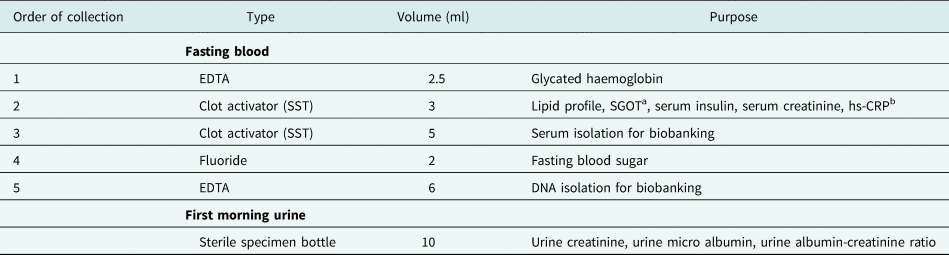
a Serum glutamic oxaloacetic transaminase.
b Highly sensitive c reactive protein.
Participants were given the options of (1) visiting the IRD; (2) visiting the main collection centre of the private hospital or one of their collection centres; or (3) have a research team visit their homes for biospecimen collection. Home visits were chosen by the majority of participants. The two teams conducting field biospecimen collection included a phlebotomist and two research assistants; one of whom had a medical background and the other in charge of driving and navigation. A single team was able to visit between 10 and 15 participants on a single day in the morning. Participants were grouped into clusters based on their geographic proximity to each other. Therefore, each participant's overnight fasting time remained between 9 and 12 h. Passive containers with plastic transport racks stored inside rigifoam boxes layered with coolant gel packs were used for specimen transportation. Biospecimens collected from the satellite centres as well as at the IRD and by the research teams were transported directly to the main laboratory of the private hospital within the strict 4-h time frame.
Processing of biospecimens for clinical investigations and biobanking
Blood and urine samples intended for clinical investigations were processed by staff at the main laboratory. Blood and urine not used in clinical investigations was discarded. The 5 ml clot activator tubes were centrifuged and the separated serum (approximately 1.5–2 ml) was immediately divided into two equal aliquots. These were transferred into screw-top cryogenic vials using micropipettes and stored in a −80 °C freezer until further testing.
Guidelines for biobanking standards have not been developed for Sri Lanka as yet. Therefore, we adopted several international guidelines to ensure integrity and viability of DNA samples. DNA extraction from the whole blood samples was carried out at IRD's genetics laboratory using a Flexi Gene 250 DNA Purification Kit (Qiagen, Germany) under sterile conditions. DNA of each participant was divided into two equal aliquots and stored in 1.4 ml cryo-vials. The paired aliquots were stored in separate cryo-boxes and transferred back to the −80 °C freezer.
The quality/purity of the extracted DNA were controlled by measuring the absorbance (A) of every sample at 260, 280, and 230 nm spectrophotometrically and determining the A 260/A 280 and A 260/A 230 ratios. For samples of good quality, A 260/A 280 is expected to lie in the range of 1.6–2.1. Lower values may indicate protein contamination whereas higher values may indicate a high content of RNA. At SLTR-b, the average ratio A 260/A 280 in general is found to be 1.8 (±0.2). The A 260/A 230 ratio was used as a secondary measure of DNA purity. Expected A 260/A 230 values for pure DNA are commonly within the range between 2.0 and 2.2. If the ratio is lower than expected, it may indicate the presence of contaminants such as proteins, Guanidine HCL (used for DNA extraction), EDTA, salts, or phenols. At SLTR-b, the average A 260/A 230 ratio is found to be 2.0 (±0.2). Integrity of DNA samples was assessed by agarose gel electrophoresis.
Clinical investigation reports
Original reports for clinical investigations were sent to participants whereas a digital copy was securely stored at IRD. We provided a cover letter which matched the participant's name to the identification number in the investigation reports. Body mass index (BMI), waist circumference, and blood pressure were included in this letter.
Biospecimen storage
Biobanking samples were labelled using cryogenic labels and stored in a large capacity −80 °C Forma 88600V freezer (Thermo Fisher Scientific, USA). A strictly controlled temperature is maintained to avoid unnecessary freeze–thaw cycles which may affect the levels of serum biomarkers [Reference Tworoger and Hankinson10] or yield of extracted genomic DNA [Reference Madisen, Hoar, Holroyd, Holroyd, Crisp and Hodes11]. During the biospecimen collection period, the freezer was initially stored in the main laboratory, which had backup generators and 24-h surveillance. The freezer was locked, and contents only accessible to authorised personnel of the COTASS 2 team.
In 2017, the −80 °C freezer and its content were transferred to the IRD's genetics laboratory while ensuring reliable power backup and greater security. A generator powered backup system was installed at IRD premises to ensure an uninterrupted power supply. It was coupled with a GSM power failure alarming system to alert the function of the power backup system using mobile short messenger service notifications.
Ethics and governance
COTASS 2 received ethical approval from the Faculty of Medical Sciences University of Sri Jayewardenepura, Sri Lanka Ethical Review Committee (reference number: 596/11), and the Psychiatry, Nursing & Midwifery Research Ethics Subcommittee, King's College London, UK (reference number: PNM/10/11-124). Data collection procedures and storage of biospecimens were in accordance with ethical standards of both institutional review boards, the ethical policies and practices of the IRD [Reference Sumathipala and Siribaddana12], and with the Helsinki Declaration of 1975. Informed written consent from all participants were obtained using two sections in the same form; one for each of the components of the main study including clinical investigations, and another section specifically designed for long-term storage of serum and DNA. Participants were over 18 years of age and could opt out of any or all of the components of COTASS 2. Participants were able to consent to (1) clinical investigations only, (2) DNA and serum storage for biobanking only, or (3) both clinical investigations and biobanking. Respondents visiting the IRD for biospecimen collection were provided with a simple meal to break their fast after sample collection. The main laboratory of the private hospital was provided with only the participants’ unique identification number, age, and gender to maintain anonymity and confidentiality. Participants with clinical investigations indicating any departures from the norm were advised to seek medical attention from the health centre nearest to them. The wide range of clinical investigations were a great benefit to many, who were attending diabetic or cardiovascular disease clinics. Ethical considerations for stored biospecimens are mentioned in the discussion section.
Results
Biospecimen collection was completed by the end of December 2014. DNA extractions were completed in February 2016, following which quality and integrity analysis of these samples continued for another 6 months and concluded in August 2016. Samples for biobanking and clinical investigations were collected from 3357 and 3464 participants, respectively. The high-participation rate was due to the availability of house visits and high standard of participant engagement by the FRAs.
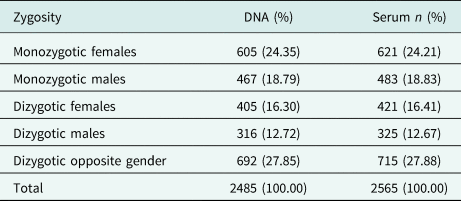
There is a slight preponderance of females in the cohort. Participant ages range from 19 to 91 years (mean age: 42.81 years). The large majority of participants were of Sinhalese ethnicity (92.94%). There was a high participation from urban areas, and many respondents were unemployed and non-manual or skilled manual workers. One percent of respondents had no education, and the many participants (46%) had been schooled up to General Certificate of Education (Ordinary Level). The COTASS 2 sociodemographic details of participants are detailed in Table 2. Seventy-four percent of the participants are individual twins, as one singleton was recruited for every pair of twins in COTASS 1. The total overall number of individual twins for DNA and serum are 2485 and 2565 respectively, and 27% are opposite gender twins. Zygosity characteristics are presented in Table 3.
Table 2. Sociodemographic characteristics of COTASS 2 participants in the SLTR-b
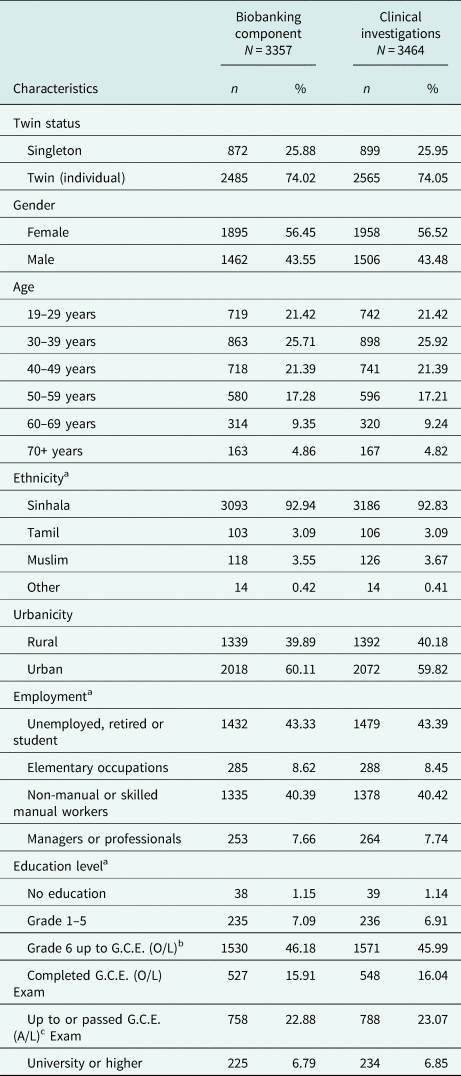
a Contains missing data.
b General Certificate of Education (ordinary level).
c General Certificate of Education G.C.E. (advanced level).
Table 3. Zygosity characteristics of twins (complete and incomplete pairs) of the SLTR-b
Anthropometric measures, blood pressure, clinical investigations, and longitudinal survey data are linked to the SLTR-b. Other data include heart rate variability data and actigraphy data from two sub-studies within COTASS 2. Table 4 provides details of all linked data, and Table 5 provides a list of selected COTASS 2 mental and physical health characteristics of participants in the SLTR-b and associations or mean differences between genders.
Table 4. Types of data linked to the SLTR-b from the longitudinal COTASS study

a World Health Organisation – Composite International Diagnostic Interview.
b The Dutch Eating Behaviour Questionnaire and the Three-Factor Eating Questionnaire were adopted for this eating disorders questionnaire.
c A questionnaire to measure the frequency of consumption of a comprehensive list of food groups in Sri Lanka.
d Recordings done on a Schiller Medilog AR12 Plus Holter Recorder at 1000 Hz.
e Data collected using Phillips Actiwatch Spectrum.
ϕ Data from twins only.
♂ Data from males only.
Table 5. Selected COTASS 2 health characteristics linked to the DNA in the SLTR-b and associations or mean differences between genders

Discussion
‘Research infrastructure is an indispensable component of research enterprise’ [13]. New knowledge and innovation can only emerge from high-quality and accessible research infrastructures which include data resources, archives, and biobanks [Reference Sumathipala and Okpaku14]. Biobanks are a valuable resource for creating advancements in science through cutting-edge omics research. However, there is a scarcity of biobanks in the South Asian region. This may be due to a multitude of reasons including limitations in infrastructure, capacity, and sociological or ethical issues less common in Western settings. In addition, the lack of an overarching research culture in developing countries may be a systematic barrier, resulting in limited new knowledge output and capacity building within low- and middle-income countries. One of the main aims of setting up the IRD was to overcome this by helping to create an overarching research culture in Sri Lanka, and providing resources and infrastructure to local and international scientists.
COTASS 1 data have been shared for higher degree programmes locally and internationally. Until now, some COTASS 2 data have already been pooled into the CODAtwins project which is the largest international effort utilising the classical twin design [Reference Silventoinen, Jelenkovic, Sund, Honda, Aaltonen, Yokoyama, Tarnoki, Tarnoki, Ning, Ji, Pang, Ordoñana, Sánchez-Romera, Colodro-Conde, Burt, Klump, Medland, Montgomery, Kandler, McAdams, Eley, Gregory, Saudino, Dubois, Boivin, Haworth, Plomin, Öncel, Aliev, Stazi, Fagnani, D'Ippolito, Craig, Saffery, Siribaddana, Hotopf, Sumathipala, Spector, Mangino, Lachance, Gatz, Butler, Bayasgala, Narandalai, Freitas, Maia, Harden, Tucker-Drob, Christensen, Skytthe, Kyvik, Hong, Chong, Derom, Vlietinck, Loos, Cozen, Hwang, Mack, He, Ding, Chang, Silberg, Eaves, Maes, Cutler, Hopper, Aujard, Magnusson, Pedersen, Aslan, Song, Yang, Lee, Baker, Tuvblad, Bjerregaard-Andersen, Beck-Nielsen, Sodemann, Heikkilä, Tan, Zhang, Swan, Krasnow, Jang, Knafo-Noam, Mankuta, Abramson, Lichtenstein, Krueger, McGue, Pahlen, Tynelius, Duncan, Buchwald, Corley, Huibregtse, Nelson, Whitfield, Franz, Kremen, Lyons, Ooki, Brandt, Nilsen, Inui, Watanabe, Bartels, van Beijsterveldt, Wardle, Llewellyn, Fisher, Rebato, Martin, Iwatani, Hayakawa, Rasmussen, Sung, Harris, Willemsen, Busjahn, Goldberg, Boomsma, Hur, Sørensen and Kaprio15]. Similarly, the newly established biobank is open for collaborative studies with academic institutions after research protocols have been accepted by the SLTR-b steering and ethics committees.
Human biospecimen collection for research and genetic studies are extremely sensitive matters especially in the developing world [Reference Cambon-Thomsen16–Reference Moodley, Sibanda, February and and Rossouw19]. Unethical collection and exportation of biospecimen during events such as the 2004 tsunami [Reference Sumathipala20] have made Sri Lankans apprehensive about biospecimen collection for research purposes. Ethical appropriateness varies between settings; understanding attitudes, views and beliefs of the community and stakeholders related to genomics research and biobanking is important [Reference de Vries, Bull, Doumbo, Ibrahim, Mercereau-Puijalon, Kwiatkowski and Parker18]. We recently completed a qualitative study on public understanding of genomic medicine and research among SLTR-b participants [Reference Chamika, Weiss, Jayaweera, Sumathipala and Ives21]. Community engagement generates public trust which is of paramount importance. The IRD was awarded grants from Wellcome Trust, UK (2009) and the Medical Research Council, UK (2018) to engage twins in research through cultural activities.
Ethics forms a crucial cornerstone in the SLTR-b. Beliefs and values of our local communities played a major role in the method and types of biospecimens collected and stored, all of which must be taken into consideration. The IRD established policies and guidelines for the Sri Lankan Twin Registry [Reference Sumathipala and Siribaddana12] which conform to accepted international standards in genomic studies. The IRD's policy on human biospecimens is to analyse the data within a national institution or laboratory if the necessary infrastructure and technology exists. To address these concerns, the IRD's genetics laboratory was established during COTASS 1 as a strategic accomplishment from an ethical and infrastructure point of view [Reference Sumathipala, Siribaddana, Hotopf, McGuffin, Glozier, Ball, Kovas, Rijsdijk, Yatawara, Pariante, Zavos, Siriwardhana, Pannala, Jayaweera, Adikari and Gunewardane22], thereby avoiding the necessity for exportation biospecimens abroad. Broad consent was not obtained from COTASS 2 participants for biobanking. Therefore, any new studies using SLTR-b specimens will require new ethical approval and consent from study participants as well.
The main strengths of the SLTR-b include the use of the twin design with a large population-based collection of DNA and serum from twins and singletons. Biospecimens are linked to a variety of clinical biomarkers, physical measures, survey data on common mental disorders, other data, and a comprehensive set of confounders. We have been able to cover a wide age range, and the younger age groups provide an opportunity to understand disease development over future follow-up studies of COTASS. Weaknesses include the lack of clinical data from COTASS 1, and limited ethnic diversity. The cohort size of the SLTR-b is relatively small in comparison with other international biobanks such as the UK Biobank [Reference Sudlow, Gallacher, Allen, Beral, Burton, Danesh, Downey, Elliott, Green, Landray, Liu, Matthews, Ong, Pell, Silman, Young, Sprosen, Peakman and and Collins23]. However, the SLTR-b was a component within COTASS 2, and therefore we believe we have done justice to the limited available funding. The biobank has limited geographic coverage (Colombo district only) and may limit generalisability to other populations.
In 2019, we received a Medical Research Council, UK grant to conduct a pilot study using the COTASS cohort (and their children) to disentangle genetic and environmental components of intergenerational risk transmission of nutritional choices on risk factors for cardio vascular disease. This is the first of its kind in a low- and middle-income country, and will use a children-of-twins design to identify the extent to which the association between parental and offspring nutrition is due to genetic and environmental transmission. The full grant is being applied for in 2020 and we shall be using DNA from the SLTR-b for GWAS to answer our research questions as well.
To conclude, the SLTR-b is a unique data-rich resource comprising genomic DNA and serum of twins and a matched sample of non-twins from Colombo, Sri Lanka. We have shown that large-scale research infrastructure development is possible in low- and middle-income countries through North–South collaborations. This is the ‘first of its kind’ biobank establishment in South Asia which will allow us to answer complex research questions, whereas aiming to address gaps in health and genetics research. It will provide opportunities for further academic collaborations local and internationally, and capacity building of future research leaders in twin and omics research. The future goal of the IRD is to create a nationally representative repository of biospecimens for use in innovative epidemiological research providing new insights into diseases burdening the region.
Acknowledgements
We would like to thank all the participants of the COTASS 2 study. We are eternally grateful to the biospecimen collection team: Amila Sandaruwan, Aruna Walisundara, Nadun Perera, Thirumalayandi Jayakumar, and all the Durdans Hospital OPD phlebotomy staff who were out and about with us even before the crack of dawn. And Dr. Jeewani Hettiarachchi who was involved in biospecimen collection at IRD. The IRD Section of Genetics team who worked on the biospecimens and the biobank: Shavindya Abeygunaratne, Rosanthy Skandhakumar, Ravindi Madanayake, Thathmi Wathsara and Charuni Rajapaksha, Nimanie Hapuarachchi, and Ruwini Cooray. We would also like thank and acknowledge Upul Tudawe and the staff of Durdans hospital: Dr. Sunil Navaratinaraja, Chandana Jayasinghe, Yasanthi Warusavithana, Vajira Jayasekara, and the medical laboratory technicians involved in sample analysis. Last but not least, we would like to thank our staff at the IRD who were involved in COTASS 2: Maneesha Jayaweera, Chamali Jayasinghe, Mekala Narangoda, Janani Marasinghe, Udeni Samanmali, Nilanthi Priyadharshani, Hansika Gamage, Pavithra Lakmali, Chavika Samarasinghe, and Vimukthi Jayasundara.
Financial support
The Colombo Twin and Singleton Follow-up Study was supported by the Wellcome Trust, UK (grant number: 093206/Z/10/Z). The Sleep Sub-study for which actigraphy data were collected was led by NG and supported by the NHMRC of Australia (grant numbers: 566529 and 571421).
Conflict of interest
Prof. Matthew Hotopf is the Principal Investigator of RADAR-CNS jointly funded the European Commission, the European Federation of Pharmaceutical Industries and Associations; includes research grants from Janssen, UCB, Merck, Biogen, and Lundbeck.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.







