Implications
Understanding the characteristics, mechanisms and principles underlying short- and long-term microbial colonization inside the digestive tract is essential to the success of microbiome-based therapies using beneficial microorganisms. This paper discusses for the first time the possibility of having indigenous Saccharomyces in the gut of livestock, a topic with important implications for health, nutrition and productivity traits. This work may prove useful to a wide variety of scientists, nutraceutical companies and livestock producers.
Introduction
Livestock have and will long be crucial for the welfare of mankind. All livestock species harbour complex microbial communities in their skin, body fluids and internal organs that support vital biochemical processes that, in turn, sustain health and productivity. Of all the microbial niches in an animal’s body, the digestive tract has attracted the most attention, in part due to the role of feed digestion for maintenance, growth and health as well as the relative ease of sampling. While animal productivity (e.g. feed efficiency) is undeniably dependent on the animal’s genetics and its relationship with the environment, we now know that this phenomenon is also closely dependent on an adequate balance between the host and its associated microbial communities, especially the gut microbiota. The relationship between the gut microbiota and feed efficiency has been demonstrated in several livestock animal species such as cattle (Guan et al., Reference Guan, Nkrumah, Basarab and Moore2008), sheep (Perea et al., Reference Perea, Perz, Olivo, Williams, Lachman, Ishaq, Thomson and Yeoman2017) and pigs (Yang et al., Reference Yang, Huang, Fang, He, Zhao, Wu, Yang, Zhang, Chen and Huang2017), thus suggesting this may be a generalized phenomenon across the whole animal kingdom. This thought is not unfounded, and decades of research on the human gut microbiota have also demonstrated a strong and dynamic interrelationship between dietary components, the gut microbiota and host’s physiology (Barratt et al., Reference Barratt, Lebrilla, Shapiro and Gordon2017).
Just as in the case of human beings, the discovery that an animal harbours a complex microbiota inside the digestive tract invariably led to the belief that this ecosystem can be modified to promote or even restore health. In livestock operations, any health improvement can have significant economic implications. One approach to modify the digestive microbiota is to use live microorganisms that when administered in adequate amounts confer a benefit to the host (also known as probiotics or direct-fed microbials (DFMs) in Animal Science, a term used since the 1990s). Understanding the principles that regulate short- and long-term microbial colonization inside the digestive tract is essential to the success of microbiome-based therapies using beneficial microorganisms (Walter et al., Reference Walter, Maldonado-Gomez and Martinez2018), especially in livestock operations. This current review focuses on one type of such beneficial microorganisms, the Saccharomyces, a genus of yeast (Figure 1).
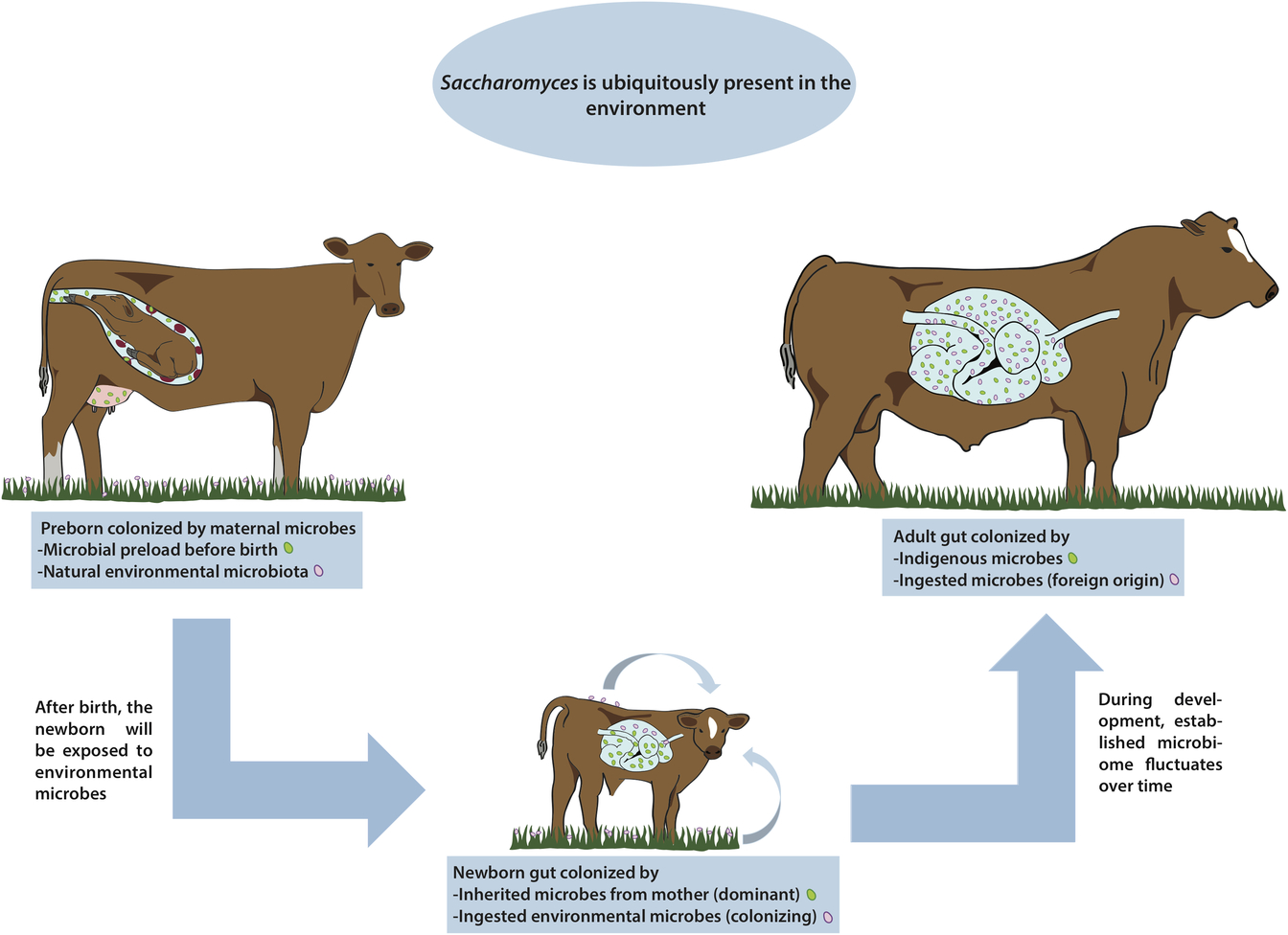
Figure 1 Are there indigenous Saccharomyces in the gut of livestock? Saccharomyces has been called a nomadic yeast due to its ubiquitous presence in many different environments. During pregnancy, animals may be colonized with microorganisms coming from the mother. Regardless of any possible prenatal microbial colonization, once the animal is born the gut is immediately colonized by microbes from the immediate surroundings, including wild Saccharomyces, and these microbes fluctuate widely over time. By the time the animal reaches adulthood, the gut is colonized by a complex microbiome comprising a combination of both indigenous and ingested microorganisms. In this paper, we discuss the presence of any truly indigenous Saccharomyces inside the cattle’s digestive system, a topic of great relevance for health, nutrition and productivity in livestock.
What is Saccharomyces?
Saccharomyces (‘sugar mushroom’, from Greek) is a member of the fungus kingdom within the eukarya domain that has been present on Earth for at least 400 million years. There are currently nine recognized species of Saccharomyces (Naseeb et al., Reference Naseeb, James, Alsammar, Michaels, Gini, Nueno-Palop, Bond, McGhie, Roberts and Delneri2017), all of which are fermentative, though focus has mostly centred on Saccharomyces cerevisiae and S. boulardii. Some nutritional limitations can cause Saccharomyces cells to enter the stationary phase, alter their morphology or sporulate (Neiman, Reference Neiman2011). Today, Saccharomyces is one of the most widely studied organisms in science due in part to their fast growth and ability to ferment a variety of carbohydrates prolifically. Saccharomyces has been examined from many different perspectives, from evolutionary genomics to biofilm biology to medical aspects in humans and animals. Saccharomyces has also received much attention because of its importance in the brewing and bakery industries. In this regard, the fermentative capacity of some yeasts and other microorganisms has been known at least since the Neolithic age, even though people did not know the mechanisms or the agents (physical or biological) that were involved in the process. According to the historical account by Boulton and Quain (Reference Boulton, Quain, Boulton and Quain2001), even as late as 1810, scientists believed that oxygen was the active ingredient that initiated fermentation by reacting with the sugary liquid. Some years later in the 1830s, Cagniard-Latour and others examined yeasts under microscope (most likely the strains of Saccharomyces) and noted that the cells proliferated by the formation of buds, and they concluded that this was evidence of life (Boulton and Quain, Reference Boulton, Quain, Boulton and Quain2001). Work by Louis Pasteur in the 1870s and by Emil Christian Hansen in the 1880s further expanded our knowledge of the fermentation process and the understanding that fermentation was indeed a microbial process. Note that fermentation is generally known to be a strictly anaerobic process, but it can be carried out at the same time as respiration, depending on the sugar levels and the particular food (e.g. bread, pickles) or drink (e.g. wine, beer) being produced.
For many years, Saccharomyces was thought to be adapted to a specific niche, that is, fruits; however, this supposition has recently been challenged by the fact that Saccharomyces can be found in many other environments (Goddard and Greig, Reference Goddard and Greig2015). For example, members of Saccharomyces have been found in skin, digestive contents, reproductive tracts, milk, plants, insects, fruits, soil, tree barks and even aquatic environments. Interestingly, wherever found, Saccharomyces is often low in abundance (Goddard and Greig, Reference Goddard and Greig2015); and growing evidence suggests that rare microbes may be the overlooked keystone species regulating the functioning of different microbial environments including host associated (Jousset et al., Reference Jousset, Bienhold, Chatzinotas, Gallien, Gobet, Kurm, Küsel, Rillig, Rivett, Salles, van der Heijden, Youssef, Zhang, Wei and Gera Hol2017).
Saccharomyces in livestock operations
Saccharomyces is one of the most widely used and studied DFMs in livestock, particularly in cattle. Based on our experience and the available literature, there is no strong rationale behind the preferred use of Saccharomyces in animal nutrition beyond the fact that S. boulardii was isolated almost one century ago and ever since has been shown to be effective to restore health in human beings (in other words, people may have been influenced by the literature in human beings to start considering the use of Saccharomyces in animals’ diets). Another possible factor was the availability of feed materials such as beer fermentation remnants. Different strains of Saccharomyces have been used to improve health, nutrition and productivity traits in most livestock animal species, from cattle to chickens, with some studies reporting measurable benefits while other studies showing no effects. This variation among the different studies can be explained by host-associated factors (e.g. age, breed, sex), environmental factors (e.g. dose of probiotics, facilities, weather), technical factors (culture v. culture-independent methods, PCR primers, genetic region to be analysed) and other unknown factors (e.g. subclinical disease, interaction between nutrients). In addition, administration of the same type of microorganism to different animal species is interesting because of the inherent anatomical and physiological differences, the differences in gut microbiota composition and metabolic activity, the different stressors that animals experience, the different pathogens affecting them and the different life productive cycles. Nonetheless, there is sufficient evidence to affirm that at least some strains of Saccharomyces can have a beneficial impact on several livestock operations in certain circumstances (EFSA Panel on Additives and Products or Substances used in Animal Feed, 2014; Alugongo et al., Reference Alugongo, Xiao, Wu, Li, Wang and Cao2017).
In vivo mode of action of Saccharomyces in livestock
As it will become clear throughout the manuscript, a brief discussion about the mode of action of Saccharomyces is useful in a context of the possible existence of autochthonous Saccharomyces. The interest in the mode of action of Saccharomyces and other beneficial microorganisms inside the animal is not only of academic merit. As mentioned above, understanding the mechanisms explaining microbial colonization inside the digestive tract is essential to the success of microbiome-based therapies using beneficial microorganisms (Walter et al., Reference Walter, Maldonado-Gomez and Martinez2018). This is particularly relevant in livestock operations, where the addition of any dietary supplement represents a cost that preferably generates an added value to the final product. Currently, two main Saccharomyces products exist in the market, fermentation products (also known as yeast cultures that supposedly do not contain viable organisms) and products containing truly live (a.k.a active dry) yeasts. This is not a trivial topic, because some commercial products containing only yeast cultures are mislabelled as live yeasts (Poppy et al., Reference Poppy, Rabiee, Lean, Sanchez, Dorton and Morley2012); and though each type of product is known to have different mechanisms of action in vivo, some (but not all, see Fonty and Chaucheyras-Durand, Reference Fonty and Chaucheyras-Durand2006) researchers have suggested that the two products may not have significant differences in their mode of action, at least in vitro (Lynch and Martin, Reference Lynch and Martin2002; Alugongo et al., Reference Alugongo, Xiao, Wu, Li, Wang and Cao2017). However, this topic is outside the scope of this manuscript; and here, when we refer to consumption of Saccharomyces, we assume that it is consumption of products containing live yeasts.
The American Type Culture Collection contains information about thousands of strains of Saccharomyces (including S. cerevisiae) and recommends to grow the organisms as ‘typical aerobic’ at 24°C to 30°C, depending on the strain. Nonetheless, Saccharomyces can grow at a wide range of temperatures, depending on the strain (Salvadó et al., Reference Salvadó, Arroyo-López, Guillamón, Salazar, Querol and Barrio2011), and is one of the few yeasts that can grow rapidly under anaerobic conditions (Ishtar Snoek and Yde Steensma, Reference Ishtar Snoek and Yde Steensma2007). This is interesting because the rumen (~39°C) is an anaerobic chamber, but considerable concentrations of dissolved oxygen enter into the digestive tract during the daily feeding cycle, especially in the liquid phase. The in vivo mode of action of Saccharomyces inside the rumen is thought to be related to the respiratory activity of the yeast (Newbold et al., Reference Newbold, Wallace and McIntosh1996), a phenomenon that requires the yeast cells to be alive and metabolically active. This is supposedly done by consuming some of the oxygen in ruminal fluid, thus preventing toxicity to the ruminal bacterial and other strict anaerobes and allowing a better performance of these organisms on feed degradation (Newbold et al., Reference Newbold, Wallace and McIntosh1996). The idea of enhancement of bacterial and fungal colonization of fibrous materials due to a diminished amount of oxygen has lasted until now (Chaucheyras-Durand et al., Reference Chaucheyras-Durand, Ameilbonne, Bichat, Mosoni, Ossa and Forano2016). Other explanations for the in vivo mode of action is nutritional competition with autochthonous ruminal microbial species as well as the supply of nutrients (e.g. amino acids, peptides, vitamins and organic acids) with the capacity to stimulate the growth of some rumen bacteria and anaerobic fungi (Fonty and Chaucheyras-Durand, Reference Fonty and Chaucheyras-Durand2006). Although we could not find any paper in the literature suggesting or demonstrating that commercial Saccharomyces can use more than one mechanism of action once inside the digestive tract or have the potential of using both aerobic and anaerobic pathways in vivo, these are not unrealistic suppositions.
Are there any indigenous Saccharomyces in the gut of livestock?
Before continuing with this review, we think it is important to clarify the use of terms regarding the origin of the microbes that are associated with a particular environment (e.g. the animal’s gut). Microbes may be of foreign origin (e.g. coming from the diet) and thus be only transiently present in the gut (the term allochthonous has been used to refer to these foreign microorganisms). On the other hand, there are microbes that are early or pioneer colonizers of a particular gut section (e.g. rumen) due to early acquisition prior to birth, during the birth process or from the immediate environment (e.g. mother) after birth (Yeoman et al., Reference Yeoman, Ishaq, Bichi, Olivo, Lowe and Aldridge2018). Several terms have been used to name these microbes such as native, autochthonous or indigenous microbes. Although we acknowledge that this is a matter of controversy, especially in light of a growing body of research on the ‘core’ microbiome, for our purposes here we will call these microbes indigenous, because in practice it is very difficult to truly define the origin of any microbe present in the gut.
Humans and animals were long considered to be born sterile (i.e. without any microorganism), but new evidence strongly suggests that newborns are preloaded with microbes and that this phenomenon begins in utero (Jami et al., Reference Jami, Israel, Kotser and Mizrahi2013; Chong et al., Reference Chong, Bloomfield and O’Sullivan2018). However, a recent paper has challenged this in ruminants due to placental architecture (Malmuthuge and Griebel, Reference Malmuthuge and Griebel2018). Regardless, the presence of microorganisms in the adult animal’s gut (particularly bacteria) is implicitly regarded as normal or indigenous microbiota (i.e. not a dietary contaminant). Efforts have been made to find a ‘core’ microbiome (i.e. organisms common across microbiomes) (Henderson et al., Reference Henderson, Cox, Ganesh, Jonker, Young and Janssen2015) based on the assumption that it plays a key role in ecosystem function, but whether this core represents truly indigenous communities has barely been mentioned in the literature (Shade and Handelsman, Reference Shade and Handelsman2012). Interestingly, growing evidence suggests the existence of an indigenous mycobiome in human beings (Nash et al., Reference Nash, Auchtung, Wong, Smith, Gesell, Ross, Stewart, Metcalf, Muzny, Gibbs, Ajami and Petrosino2017). If true, then it is likely that animals also possess an indigenous mycobiome. However, several authors have mentioned that Saccharomyces is not indigenous to the human or animal digestive tract, a belief that is not necessarily held for all the other members of the gut microbiota, especially bacteria and also other fungi, even though the colonization challenges may be similar for different microorganisms. In fact, to our knowledge, no one has ever suggested the possible existence of indigenous Saccharomyces in the gut, either in humans or in animals. However, different anatomical locations in the digestive tract harbour various bacterial communities, as do co-located digesta or mucosal-associated microenvironments (Yeoman et al., Reference Yeoman, Ishaq, Bichi, Olivo, Lowe and Aldridge2018). It is reasonable to assume this is also true for fungi. This is important because most studies still rely on luminal contents only (mostly faeces); therefore, it is likely that our knowledge of truly indigenous (or truly transient) microbes is strongly methodologically biased. This is also important because, for example, feed efficiency may be related to microorganisms that are present in other sections of the digestive system and thus not necessarily present in abundance in faeces (Perea et al., Reference Perea, Perz, Olivo, Williams, Lachman, Ishaq, Thomson and Yeoman2017).
In an early descriptive study of yeasts in the bovine caecum, van Uden and Do Sousa (Reference van Uden and Do Sousa1957) mentioned that some yeast species (such as S. cerevisiae, which was actually found in the samples analysed) are not true inhabitants of the digestive tract of human and other animals, without explaining the reasons behind their thought, perhaps reflecting the contemporary view of the first half of the 20th century. Williams et al. (Reference Williams, Tait, Innes and Newbold1991) also mentioned that S. cerevisiae is not normally encountered in the rumen and cited this as a reason that exogenous supplementation with these organisms would improve digestion in cattle. In another review, Wallace (Reference Wallace1994) mentioned that S. cerevisiae is not part of the indigenous ruminal species; therefore, S. cerevisiae could be a more appropriate vehicle than indigenous ruminal organisms for implementing the benefits of recombinant DNA technology in the area of ruminal nutrition and fermentation. Although this topic has been ignored ever since, the thought that Saccharomyces is not indigenous to the gut of livestock (or even to the whole group of mammals, see Martins et al., Reference Martins, Nardi, Arantes, Rosa, Neves and Nicoli2005) has endured until now (Fonty and Chaucheyras-Durand, Reference Fonty and Chaucheyras-Durand2006).
Considering the fact that cattle and other animals have been continuously exposed to different environmental Saccharomyces over millions of years, and the generalized belief that the presence of Saccharomyces in the gut only reflects dietary contamination (see above), an explanation of why Saccharomyces never became indigenous is necessary. The load of Saccharomyces and other microorganisms in animal feed is a very interesting topic but unfortunately has not been well studied (Santos et al., Reference Santos, Golt, Joerger, Mechor, Mourão and Kung2017). We hypothesize that the Saccharomyces present in the feed may have never become adapted to the gut because every time the animal feeds, it feeds with different types of Saccharomyces; therefore by the time one strain attempts to colonize, it gets removed (out-competed) by other strains. While this may make sense, given the high heterogeneity among the different types of Saccharomyces (Strope et al., Reference Strope, Skelly, Kozmin, Mahadevan, Stone, Magwene, Dietrich and McCusker2015), the thought is somehow in conflict with the famous like-will-to-like concept, though researchers acknowledge the possibility that this concept is likely not universal for all microbes in all circumstances (Winter and Bäumler, Reference Winter and Bäumler2014). It is also important to note that at least S. cerevisiae is often present low in abundance compared to other microbes (Goddard and Greig, Reference Goddard and Greig2015); therefore, it is also feasible to hypothesize that at least some strains of Saccharomyces may simply not have the natural capacity (or need) to massively colonize any particular environment. However, as mentioned above, rare and low prevalent microbes are both very common in different ecosystems and may play very important roles (Jousset et al., Reference Jousset, Bienhold, Chatzinotas, Gallien, Gobet, Kurm, Küsel, Rillig, Rivett, Salles, van der Heijden, Youssef, Zhang, Wei and Gera Hol2017), yet they are often ignored or removed from DNA sequencing analyses.
If we accept the possibility that there are no truly indigenous Saccharomyces in the animals’ gut, why are they effective in vivo? While it has been proposed that the existence of related microbial species may help incoming foreign organisms to get established (like-will-to-like concept; Winter and Bäumler, Reference Winter and Bäumler2014), the same authors propose that the rules behind this phenomenon likely do not apply to all microbes alike. Also, others have suggested that intestinal colonization by probiotic bacteria may be higher in subjects having lower concentrations of these bacteria before ingestion of probiotics (Vitali et al., Reference Vitali, Ndagijimana, Cruciani, Carnevali, Candela, Guerzoni and Brigidi2010) and the engraftment of one strain of Bifidobacterium longum was associated with less occurrence of indigenous B. longum (Maldonado-Gómez et al., Reference Maldonado-Gómez, Martínez, Bottacini, O’Callaghan, Ventura, van Sinderen, Hillmann, Vangay, Knights, Hutkins and Walter2016). Yet this theory would only apply if Saccharomyces was truly indigenous, or whether other species of Saccharomyces, which might offer competition to the DFM, was linked to successful colonization, neither of which has been definitively shown. It is also likely that the dynamics for bacterial colonization differs from that of fungi.
Here we raise the possibility that Saccharomyces is effective because it is a foreign organism in cattle and finds little competition at least with regard to oxygen consumption. In this regard, abundant DFM Saccharomyces cerevisiae was found in epithelial or solid-associated fractions in the rumen (Ishaq et al., Reference Ishaq, AlZahal, Walker and McBride2017) near the top where oxygen content is higher. However, little is known about aerobic organisms in the gut of livestock. In the rumen, aerobic and facultative anaerobic bacteria are abundant early in life and then decline over time (Jami et al., Reference Jami, Israel, Kotser and Mizrahi2013). Saccharomyces is often fed to mitigate the likelihood of acidosis from feeding a high-grain diet (e.g. Ishaq et al., Reference Ishaq, AlZahal, Walker and McBride2017); the combination of the two may provide access to fermentative sugars that support Saccharomyces by reducing competition for nutrients. Similarly, consumption of fruits along with the associated Saccharomyces might allow for colonization and benefits in humans. Regardless, we acknowledge the efforts in the field of microbiome modulation with live microbes and the challenges associated with the analysis of microbial engraftment within the gut (Walter et al., Reference Walter, Maldonado-Gomez and Martinez2018).
Mode of action in vivo of Saccharomyces in human beings
It is also important for this work to briefly touch on the human microbiota because the mode of action in vivo of Saccharomyces (i.e. S. boulardii) in humans has not been related to oxygen consumption by any author to date and also because the human microbiota has been much better studied compared to the animal microbiota.
Saccharomyces boulardii has proved its effectiveness in preventive and therapeutic treatment of many diseases of the human digestive tract. Saccharomyces boulardii was discovered by Henri Boulard in the 1920s after having observed that some indigenous peoples in Indo-China consumed the skins of various tropical fruits as an antidiarrheal medication. Given the source (e.g. fruit’s skin) of the original S. boulardii strain, it can be argued that at least this strain is not an obligate anaerobe and is definitely not indigenous to the human or animal gut, yet there is enough evidence to suggest that this (foreign) organism can have a significant effect on human health; however, questions remain about its safety (Roy et al., Reference Roy, Jessani, Rudramurthy, Gopalakrishnan, Dutta, Chakravarty, Jillwin and Chakrabarti2017) as in the case of S. cerevisiae (Strope et al., Reference Strope, Skelly, Kozmin, Mahadevan, Stone, Magwene, Dietrich and McCusker2015). The risk of disease is not exclusive to consumption of Saccharomyces or any other particular probiotic or combination of probiotics either in humans or in animals. In other words, any ingested microorganisms may cause disease if it encounters the appropriate conditions to do so, for example, a subject that is immunocompromised. The use and safety of Saccharomyces and other microorganisms for animal feed is regulated as a food additive (e.g. by the Food and Drug Administration in the United States and the European Food Safety Authority in the European Union, see EFSA Panel on Additives and Products or Substances used in Animal Feed, 2012). Safety aspects that have been discussed in this regard include physical state of the final product and antibiotic sensitivity; other safety issues related to probiotic administration in livestock operations are not well understood due to the fact that many livestock operations measure health indirectly by means of measuring productivity traits (e.g. milk production).
The beneficial effect of S. boulardii in human health has been hypothesized to involve several mechanisms, including preservation of intestinal barrier function, anti-inflammatory ability, pathogen binding as well as reduction in apoptosis of and adhesion to intestinal epithelial cells (McFarland, Reference McFarland2010; Stier and Bischoff, Reference Stier and Bischoff2016). Some of these effects are mediated by a direct physical interaction between the yeasts and the intestinal brush border membranes (Buts and De Keyser, Reference Buts and De Keyser2010). Interestingly, the interaction of S. boulardii with a healthy uninflamed intestinal mucosa is limited (i.e. small systemic humoral immune response, induction of only few transcriptional changes), at least in mice (Hudson et al., Reference Hudson, McDermott, Stewart, Hudson, Rios, Fasken, Corbett and Lamb2016). To our knowledge, the effects of in vivo S. boulardii in humans have not been related to their respiratory activity as in the case of S. cerevisiae in ruminants. The question of mode of action deserves more attention, given that similar organisms (Saccharomyces spp.) are offered to animals with highly different anatomical and physiological differences, including horses, cattle, chickens, pigs and even aquatic animal species.
Are there any indigenous Saccharomyces in the gut of human beings?
Saccharomyces is an abundant (average 15% of all sequence reads) and prevalent member of the milk mycobiome in healthy mothers (Boix-Amorós et al., Reference Boix-Amorós, Martinez-Costa, Querol, Collado and Mira2017). This is very interesting because it may imply that human beings are exposed to Saccharomyces since very early in life. In this paper, the authors discussed that this may be due to skin contamination and also raise the intriguing possibility of an entero-mammary pathway involving an internal route (Boix-Amorós et al., Reference Boix-Amorós, Martinez-Costa, Querol, Collado and Mira2017). Saccharomyces has also been detected in human faeces a number of times (Nash et al., Reference Nash, Auchtung, Wong, Smith, Gesell, Ross, Stewart, Metcalf, Muzny, Gibbs, Ajami and Petrosino2017), being sometimes the dominant genus representing over half of total relative abundance (Huseyin et al., Reference Huseyin, Cabrera Rubio, O’Sullivan, Cotter and Scanlan2017). Nonetheless, the thought that the presence of Saccharomyces in the human gut may be solely due to the dietary contamination as stated by these and other authors, with the argument that ‘it is a domesticated species of fermentations whose wild niche is associated with plants’ (Hallen-Adams and Suhr, Reference Hallen-Adams and Suhr2017). Despite identifying Saccharomyces in the human gut at high levels using culture-independent techniques, Huseyin et al. (Reference Huseyin, Cabrera Rubio, O’Sullivan, Cotter and Scanlan2017) also suggested that this may be due to dietary contamination and proposed the theory that Saccharomyces cannot be native due to the temperatures of the environments from which they had been isolated. Interestingly, Seddik et al. (Reference Seddik, Ceugniez, Bendali, Cudennec and Drider2016) isolated one strain of S. cerevisiae from faeces of Algerian infants that was determined to be safe and usable as probiotic, and Rajkowska and Kunicka-Styczyńska (Reference Rajkowska and Kunicka-Styczyńska2010) investigated the probiotic properties of one strain of Saccharomyces isolated from chicken faeces (note that body temperature in chickens is slightly higher compared to mammals). Since the isolated Saccharomyces were obviously viable upon collection of faecal samples, the proposal by Huseyin et al. (Reference Huseyin, Cabrera Rubio, O’Sullivan, Cotter and Scanlan2017) that Saccharomyces cannot be indigenous due to the temperatures of the environments from which they had been isolated is likely to be incorrect (actually, McFarland (2010) mentions that the optimal temperature for growth of S. boulardii is 37°C). On top of this, Saccharomyces was not identified in a recent diet study as a food-sourced microorganism (David et al., Reference David, Maurice, Carmody, Gootenberg, Button, Wolfe, Ling, Devlin, Varma, Fischbach, Biddinger, Dutton and Turnbaugh2014), thus further questioning the possibility that its presence solely reflects dietary contamination.
Overall, researchers hold the (rather unfounded) belief that the presence of Saccharomyces in digestive contents solely reflects dietary contamination. Interestingly, however, it has been shown that S. cerevisiae decreased in faecal samples of patients with inflammatory bowel disease (Sokol et al., Reference Sokol, Leducq, Aschard, Pham, Jegou, Landman, Cohen, Liguori, Bourrier, Nion-Larmurier, Cosnes, Seksik, Langella, Skurnik, Richard and Beaugerie2017) and other disorders. If these organisms were not indigenous to the gut, why would their abundance be lower in specific health disorders? If they are not indigenous, then they have to come from the food and therefore studies by Sokol et al. (Reference Sokol, Leducq, Aschard, Pham, Jegou, Landman, Cohen, Liguori, Bourrier, Nion-Larmurier, Cosnes, Seksik, Langella, Skurnik, Richard and Beaugerie2017) would necessarily have to imply one or two options: (1) a lower capacity to stay or survive in the gut during certain health disorders (perhaps as a result of an adverse immune reaction), including a potentially diminished ability to attach to the mucosa or (2) a detachment of (indigenous?) Saccharomyces from the mucosa. Unfortunately, the authors of this paper did not discuss their findings with a perspective of indigenous or transient microbiota.
Should we choose an indigenous or a foreign microorganism as a potential probiotic?
This question is particularly relevant to nutraceutical companies aiming to develop new and more effective formulations containing beneficial microorganisms for livestock. As reviewed above, neither S. boulardii nor any other Saccharomyces used in brewing, wine making or bakery were indigenous to the human or animal gut, yet there is enough evidence that the consumption of these (foreign) microorganisms can elicit beneficial effects. Although it may sound surprising for the unfamiliar reader, this phenomenon is actually not unexpected, especially in the light of new evidence showing that bacteria from diverse habitats (including soil and estuarine microbial mats) can not only colonize but also compete inside the mouse gut (Seedorf et al., Reference Seedorf, Griffin, Ridaura, Reyes, Cheng, Rey, Smith, Simon, Scheffrahn, Woebken, Spormann, Van Treuren, Ursell, Pirrung, Robbins-Pianka, Cantarel, Lombard, Henrissat, Knight and Gordon2014). This capacity to survive in different environments has been less studied in non-bacterial organisms but is also likely true for other life forms such as members of the domains eukarya (Saccharomyces included, see Goddard and Greig, Reference Goddard and Greig2015) and archaea.
Studies of the human microbiome have shown that each individual is highly unique in terms of its associated microbiota, even at the species and strain level (The Human Microbiome Project Consortium, 2012), and this is likely true for the animal microbiota. Taking this into account, even the isolation of one truly indigenous microorganism from one or more hosts (e.g. Lactobacillus from one or more cows) is very likely not to be related to the indigenous microbiota of other hosts, even from the same animal species or the same breed. In the case of cattle and perhaps other animals as well, at the field level it is simply impossible to provide sterile feed to determine truly indigenous microbes to the host. All things considered, we would like to bring up an example that may help compile these and other thoughts into the main objective of this work.
Despite a generalized lack of information with regard to the mycobiome (indigenous or not) in pigs and other animals, different species and strains of Saccharomyces have been used in the pig industry for at least 70 years. Based on a comprehensive survey of gut mycobiome in piglets using culture methods, and supported by the notion that knowledge of the yeasts normally present in the digestive tract can help understand the effect of these organisms on the host, Urubschurov et al. (Reference Urubschurov, Janczyk, Pieper and Souffrant2008) isolated a variety of yeasts in an experimental farm, of which Kazachstania slooffiae was found to be dominant. This is an understandable and intelligent approach to start looking for potential probiotic species to work in particular circumstances (e.g. weaning). Based on their original findings, the same research group studied the effect of a K. slooffiae strain isolated from pigs’ faeces on growth performance, intestinal microbial metabolites and gut microbiota (Urubschurov et al., Reference Urubschurov, Büsing and Freyer2016). While we acknowledge that this approach may not necessarily lead to the best potential candidates as probiotic species, and that multiple in vitro tests are advisable to determine the functionality, it is also true that this line of thought (i.e. isolation of autochthonous microorganisms) is worth pursuing. Other approaches such as the use of microorganisms from soil and other environments (Seedorf et al., Reference Seedorf, Griffin, Ridaura, Reyes, Cheng, Rey, Smith, Simon, Scheffrahn, Woebken, Spormann, Van Treuren, Ursell, Pirrung, Robbins-Pianka, Cantarel, Lombard, Henrissat, Knight and Gordon2014) may also be considered to select new DFMs.
Concluding remarks
All livestock animals depend on their gut microbiota to be healthy and productive. Despite more technology to understand the role of the gut microbiota in humans and animals, there are many questions that remain unanswered with regard to Saccharomyces, particularly the two aspects: Saccharomyces ecology in nature and host–Saccharomyces relationship. For example, an intriguing role of social wasps has been suggested in S. cerevisiae ecology, reproduction and evolution (Stefanini et al., Reference Stefanini, Dapporto, Legras, Calabretta, Di Paola, De Filippo, Viola, Capretti, Polsinelli, Turillazzi and Cavalieri2012), even though it is likely that the Saccharomyces found in these and other insects are not truly indigenous. In addition, sporulation in soil has been suggested to be an overwinter survival strategy in S. cerevisiae (Knight and Goddard, Reference Knight and Goddard2016), but this phenomenon and other aspects of survival have not been deeply investigated in soil, digestive contents or any other environment. The animal–Saccharomyces relationship is relevant not only in the context of whether it is indigenous or not but also within the context of potential host specificity or the co-location in particular intestinal settings. Saccharomyces has not been found in some animal species, for example, the study of pigs discussed above (Urubschurov et al., Reference Urubschurov, Janczyk, Pieper and Souffrant2008), and one study found S. cerevisiae in three rumen fractions (i.e. rumen fluid, solids and epimural) of Holstein dairy cows but its occurrence was very low (<1%), and its occurrence was not different between animals supplemented with live Saccharomyces and those not supplemented and also between healthy cows and cows with sub-acute ruminal acidosis (Ishaq et al., Reference Ishaq, AlZahal, Walker and McBride2017). This is particularly interesting in the context of Saccharomyces because some researchers believe that this organism may exert beneficial effects mostly in the event of dysbiosis and other diseases. We hope that these and the other thoughts discussed in this work prove useful to a wide variety of scientists, nutraceutical companies and livestock producers.
Acknowledgements
None.
J. F. Garcia-Mazcorro 0000-0001-6047-4361
Declaration of interest
JFGM is an employee of MNA de México, a company of Animal Nutrition. MVRH is an employee of RT Biotech, a company that commercializes live yeast mainly for beer production.
Ethics statement
Not applicable.
Software and data repository resources
Not applicable.



