Background
The Human Genetics Society of Australasia (HGSA) supports the genetic health of the Australian and New Zealand populations. The Society advocates for the safe, ethical, and effective use of genetic information in healthcare. It promotes the establishment of high standards of professional practice, contributes to professional and lay education, and promotes public awareness of human genetics.
Historically, the focus in human genetics has been primarily on Mendelian conditions determined by a strong single gene effect. Recent developments in technology and analysis have allowed the measurement of more complex genetic effects that can be expressed in the form of polygenic scores (PGS). There has been a rapid expansion of research information in the field of PGS, albeit with ongoing debate about their readiness for implementation into healthcare, including discussion of their potential benefits and risks, and persistent gaps in the evidence (Hunter & Drazen, Reference Hunter and Drazen2019; Jia et al., Reference Jia, Lu, Wen, Long, Liu, Tao, Li, Denny, Shu and Zheng2020; Lambert et al., Reference Lambert, Abraham and Inouye2019; C. M. Lewis & Vassos, Reference Lewis and Vassos2017, Reference Lewis and Vassos2020; Palk et al., Reference Palk, Dalvie, de Vries, Martin and Stein2019; Polygenic Risk Score Task Force of the International Common Disease, 2021; Torkamani et al., Reference Torkamani, Wineinger and Topol2018; Wald & Old, Reference Wald and Old2019). PGS hold great promise to improve the health of individuals and populations.
Unlike diagnostic genetic testing, PGS provide health information in the form of an estimate of risk, which in some contexts is highly valuable. PGS are frequently included alongside other types of risk information in models that provide a combined personal estimate of disease risk. They can also be used beyond estimating risk; for example, to facilitate diagnosis and predict prognostic outcomes as well as guide therapeutic interventions.
Although PGS have begun to transition from discovery research to studies of clinical implementation in some fields, their use remains nascent. They are available to be ordered from a small number of commercial providers but have not been adopted as a component of standard practice in Australian or New Zealand health services, reflecting some important limitations with regard to their use in routine clinical care or population health programs.
This position statement outlines the HGSA’s stance on the use of disease-associated PGS in clinical practice and population health. Its focus is on health conditions rather than nondisease traits. The statement identifies and discusses current limitations with PGS and points to the additional evidence required and issues to be considered before PGS can be appropriately, safely, effectively and ethically implemented into the Australasian healthcare system and routinely used for individual patients or populations.
Box 1. Definitions
Single Nucleotide Polymorphism (SNP): a genomic variant at a single base position in the DNA. A SNP is the most common variation in the human genome. This term is often used by convention in the PGS field to also include common genomic variants that involve multiple nucleotides. Also referred to as a single nucleotide variant (SNV).
Genomewide association studies (GWAS): an observational research approach that involves genotyping a large number of variants in a substantial number of cases and controls to identify genetic variations associated with the occurrence of a particular trait or disease.
Polygenic score (PGS): a score quantifying an individuals’ genetic liability to a disorder or a trait. The score combines information about many genetic variants associated with the disease or trait, usually weighted based on the effect size from the discovery GWAS and standardized using the distribution in a relevant population. May also be referred to as a polygenic risk score, polygenic hazard score, or a genetic or genomic risk score (Wand et al., Reference Wand, Lambert, Tamburro, Iacocca, O’Sullivan, Sillari, Kullo, Rowley, Dron, Brockman, Venner, McCarthy, Antoniou, Easton, Hegele, Khera, Chatterjee, Kooperberg, Edwards and Wojcik2021; Yanes, McInerney-Leo et al., Reference Yanes, McInerney-Leo, Law and Cummings2020).
Integrated risk model: a model that combines PGS information with additional risk factors such as age, sex, clinical measurements, environmental risk factors and other biomarkers or measurements to provide a single composite risk estimate. Sometimes referred to as an integrated risk score, personalized risk score or holistic risk score (Wand et al., Reference Wand, Lambert, Tamburro, Iacocca, O’Sullivan, Sillari, Kullo, Rowley, Dron, Brockman, Venner, McCarthy, Antoniou, Easton, Hegele, Khera, Chatterjee, Kooperberg, Edwards and Wojcik2021; Yanes, McInerney-Leo et al., Reference Yanes, McInerney-Leo, Law and Cummings2020).
Clinical utility: a multidimensional concept for which there is no single definition. In clinical genetics, clinical utility can refer to the effect of genetic testing information on diagnosis, prognosis, therapeutic management, the health and psychological wellbeing of patients and their relatives, and healthcare system costs. Clinical and personal utility are interlinked (Foster et al., Reference Foster, Mulvihill and Sharp2009; Kohler et al., Reference Kohler, Turbitt and Biesecker2017; Walcott et al., Reference Walcott, Miller, Dunsmore, Lazor, Feldman and Hayeems2021).
Personal utility: although there is no single definition, personal utility can be defined as the value of the information to the person being tested. Clinical and personal utility are interlinked (Foster et al., Reference Foster, Mulvihill and Sharp2009; Kohler et al., Reference Kohler, Turbitt and Biesecker2017; Walcott et al., Reference Walcott, Miller, Dunsmore, Lazor, Feldman and Hayeems2021).
Population health: the health status of groups or whole populations, where policies and interventions aim to improve population health outcomes.
What are PGS and how are they calculated?
PGS are a measure of what can be called ‘genetic liability’ and reflect the continuous spectrum of risk found in the population. They stand, in contrast, to the rare genetic variants with a strong binary effect on risk, with the variant being present or absent, which have been the traditional focus of clinical genetics (C. M. Lewis & Vassos, Reference Lewis and Vassos2017). PGS generally provide information that can be used to enhance or guide, rather than replace, existing risk prediction models and diagnostic pathways. PGS can capture risk not obtained by other risk predictors commonly used in clinical genetics, including family history and monogenic disease variants (Jia et al., Reference Jia, Lu, Wen, Long, Liu, Tao, Li, Denny, Shu and Zheng2020). Some groups view PGS as akin to other biomarkers commonly used to assess risk, such as cholesterol, whereas others do not, primarily due to the unchanging nature of an individual’s genetic makeup (Moorthie, Reference Moorthie, Hall, Janus, Brigden, Babb de Villers, Blackburn, Johnson and Kroese2021).
Genomewide association studies (GWAS) yield summary statistics that describe the effect size and the statistical significance of the association between a variant and the outcome of interest (Visscher et al., Reference Visscher, Wray, Zhang, Sklar, McCarthy, Brown and Yang2017). These associations are combined to generate a PGS that acts as an estimate of an individual’s germline risk provided as a numerical indicator for a specific disease or trait (Torkamani et al., Reference Torkamani, Wineinger and Topol2018; Wand et al., Reference Wand, Lambert, Tamburro, Iacocca, O’Sullivan, Sillari, Kullo, Rowley, Dron, Brockman, Venner, McCarthy, Antoniou, Easton, Hegele, Khera, Chatterjee, Kooperberg, Edwards and Wojcik2021). The performance of a PGS as a predictor of disease risk (or other outcomes) is dependent, in part, on the quality and power of the GWAS that inform it and how well these studies reflect the population where the PGS is being applied, particularly with respect to genetic ancestry. As of 2021, approximately 86% of GWAS participants were of European ancestry, despite representing less than 16% of the global population (Fatumo et al., Reference Fatumo, Chikowore, Choudhury, Ayub, Martin and Kuchenbaecker2022; Martin et al., Reference Martin, Kanai, Kamatani, Okada, Neale and Daly2019). This disparity has resulted in poorer PGS performance in non-European populations. More inclusive genomic research is widely recognised as a priority by the genetics community, and various studies are now underway that aim to develop more diverse databases that are representative of global populations (Fatumo et al., Reference Fatumo, Chikowore, Choudhury, Ayub, Martin and Kuchenbaecker2022).
Many methods have been proposed to develop PGS, with the optimal approach dependent on the genetic architecture of a specific disease and the requirements of a particular clinical or population health application. The development of new and improved methods of constructing PGS (e.g., by applying new statistical techniques, incorporating functional information for variants, or by combining information across diseases and traits) is a very active area of research, with the strategies employed for variant selection and weighting of individual variants differing between diseases and populations. No consensus has yet emerged around an optimal methodology.
When considering the suitability of a PGS for clinical implementation the same standards of evidence used generally in clinical practice should be applied (Guyatt et al., Reference Guyatt, Oxman, Akl, Kunz, Vist, Brozek, Norris, Falck-Ytter, Glasziou, DeBeer, Jaeschke, Rind, Meerpohl, Dahm and Schunemann2011). Despite the common methodologies, the performance of each PGS as a risk prediction tool requires separate validation in adequately powered, independent datasets relevant to the intended implementation in order to determine key metrics, such as the score distribution, calibration, discrimination (sensitivity/specificity at different risk thresholds) and predictive ability (Choi et al., Reference Choi, Mak and O’Reilly2020). Notably, some of these aspects must be evaluated in prospective cohort studies and cannot be determined from case-control studies alone (Lambert et al., Reference Lambert, Abraham and Inouye2019; C. M. Lewis & Vassos, Reference Lewis and Vassos2020). The increasing availability of large-scale data and the proliferation of different methods has led to the development of hundreds of PGS for different conditions, including multiple cancers and cardiovascular disease, with some of them showing promising predictive performance (Lambert et al., Reference Lambert, Abraham and Inouye2019; C. M. Lewis & Vassos, Reference Lewis and Vassos2020; Wand et al., Reference Wand, Lambert, Tamburro, Iacocca, O’Sullivan, Sillari, Kullo, Rowley, Dron, Brockman, Venner, McCarthy, Antoniou, Easton, Hegele, Khera, Chatterjee, Kooperberg, Edwards and Wojcik2021).
However, a much smaller number have currently met the accepted standard of evidence required for a test to be used for clinical care outside the setting of a research study.
Current Availability of PGS in Australia and New Zealand
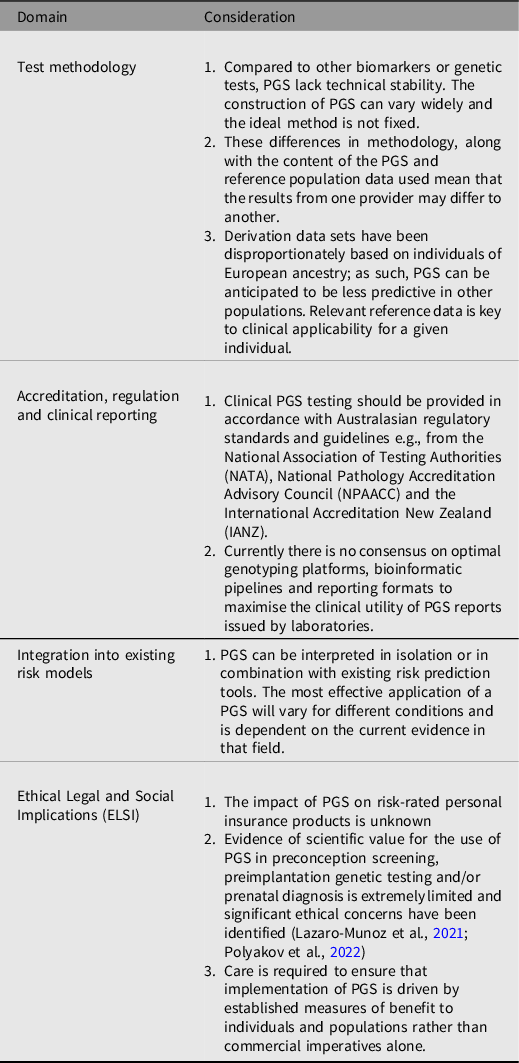
The readiness of PGS for clinical implementation remains an issue that requires further consideration. However, this statement recognises that PGS are currently available commercially for use in several clinical settings, including for more contested applications, such as preimplantation embryo screening (K. W. Davis, Reference Davis2021). Similarly, individuals are increasingly accessing PGS testing through research studies and online direct-to-consumer testing, which may lead them to seek support from their healthcare provider regarding interpretation of the results. Clinicians currently utilizing PGS in the clinical care of individuals or population health should proceed with caution as there are many caveats that deserve closer scrutiny, as outlined in Table 1. It is the professional and ethical responsibility of an ordering clinician to understand the benefits and limitations of the test and determine whether there is sufficient evidence to support its use in clinical care. Clinicians considering these tests should also understand the potential ethical, legal and social dimensions of the PGS, and as with any genetic test, the requirement for informed consent and effective communication of the implications of the results (K. W. Davis, Reference Davis2021).
Table 1. Considerations for PGS Implementation into Australasian Healthcare Systems
The Potential Clinical Application of PGS in Healthcare
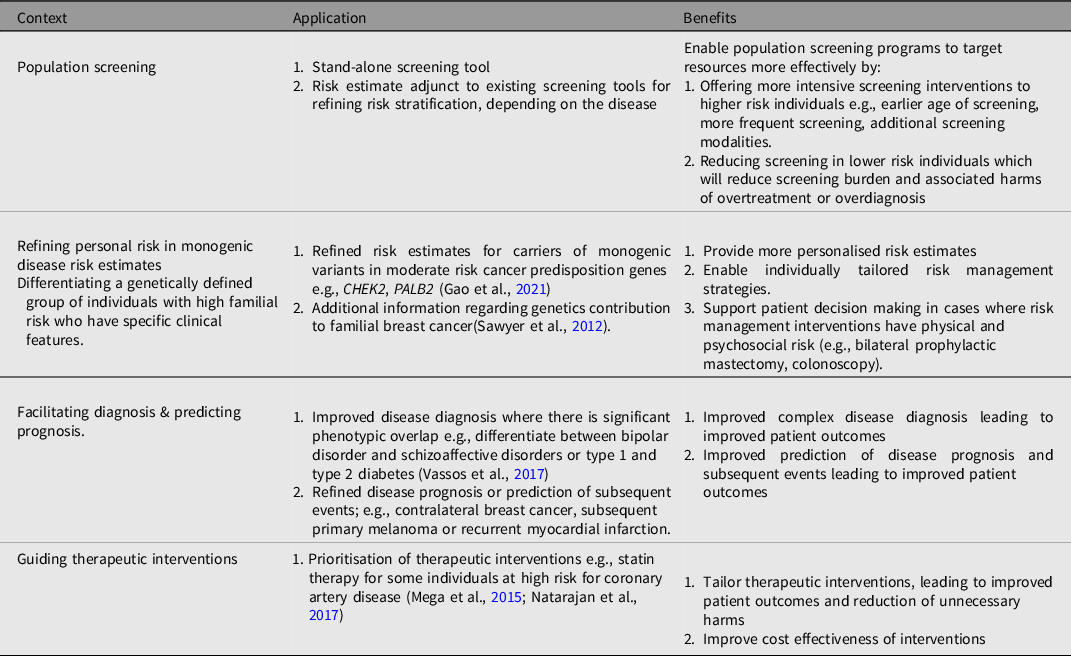
It is possible to develop a PGS to assess the risk of most common disorders and the potential clinical application of PGS is broad, but the pathway to clinical implementation is likely to be context dependent. Proposed applications of PGS (summarized in Table 2) include population screening, modifying or refining risk estimates in individuals with monogenic disease, facilitating diagnosis and predicting prognostic outcomes, as well as guiding therapeutic interventions (C. M. Lewis & Vassos, Reference Lewis and Vassos2017; Moorthie et al., Reference Moorthie, Hall, Janus, Brigden, Babb de Villers, Blackburn, Johnson and Kroese2021; Torkamani et al., Reference Torkamani, Wineinger and Topol2018).
Table 2. Examples of potential applications of PGS in healthcare
What Have We Learned to Date?
The integration of polygenic scores into clinical care and population health is likely to involve a different path to that taken for Mendelian genetics and rare conditions. It can be anticipated to require updating of several aspects of the current model of practice, described below. However, while there are many differences, there are also similarities in relation to fundamental issues already addressed in the clinical application of Mendelian genetics and which will have continuing relevance in the polygenic context. The extensive knowledge and experience gained from the development of clinical genetics in practice should continue to inform the implementation of PGS, preventing duplication of efforts and waste of resources.
Psychosocial Implications of Genetic Information
Historically, concerns have been raised about the potential for the information that arises from genetic testing in Mendelian disorders to lead to psychological harm (Kash, Reference Kash1995; Smith-Uffen et al., Reference Smith-Uffen, Bartley, Davies and Best2021). In fact, the evidence of long-term adverse psychosocial outcomes resulting from genetic testing in Mendelian disease is limited and there is now substantial contrary evidence demonstrating that information arising from genetic testing can have psychological value (Oliveri et al., Reference Oliveri, Ferrari, Manfrinati and Pravettoni2018; Ringwald et al., Reference Ringwald, Wochnowski, Bosse, Giel, Schaffeler, Zipfel and Teufel2016; Yanes et al., Reference Yanes, Willis, Meiser, Tucker and Best2019). Notwithstanding short-term distress, information arising from genetic testing can be viewed as a way for individuals to gather important information to enable proactive health management.
Concerns have now also been raised about psychosocial harms following PGS, although evidence is again limited (Fenton et al., Reference Fenton, Smit, Keogh and Cust2018; Forrest et al., Reference Forrest, Sawyer, Hallowell, James and Young2019; Wallingford et al., Reference Wallingford, Kovilpillai, Jacobs, Turbitt, Primiero, Young, Brockman, Soyer, McInerney-Leo and Yanes2022; Young et al., Reference Young, Forrest, Rasmussen, James, Mitchell, Sawyer, Reeve and Hallowell2017). Research to date demonstrates that testing for PGS offered in clinical practice or population health has a high uptake, is generally acceptable to providers and recipients, and aligns well with the general conception of heritability in the population that expects inherited features to reflect a genetic contribution from both parents (Marteau & Richards, Reference Marteau and Richards1996; Willis et al., Reference Willis, Smith, Meiser, James, Ballinger, Thomas, Yanes and Young2021; Young et al., Reference Young, Forrest, Rasmussen, James, Mitchell, Sawyer, Reeve and Hallowell2017). Studies in the clinical setting have found a good level of knowledge among patients of the broad concepts related to polygenic risk information; for example, mode of inheritance, risk for other family members (Young et al., Reference Young, Forrest, Rasmussen, James, Mitchell, Sawyer, Reeve and Hallowell2017). Acceptability may also be influenced by the personalized nature of risk information derived from PGS, which does not inform risk for family members to the same extent as Mendelian genetic testing information (Cox et al., Reference Cox, Heudel, Henry and Pivot2018); women from high-risk families have described relief about the nature of personalized risk, that is, risk for family members (Yanes, Kaur et al., Reference Yanes, Kaur, Meiser, Scheepers-Joynt, McInerny, Barlow-Stewart, Antill, Salmon, Smyth, James and Young2020). Despite these reassuring findings, some individuals have been shown to experience greater distress, especially in a setting where PGS is reported for multiple conditions or where healthcare providers are not involved in the delivery of results (Haga et al., Reference Haga, Barry, Mills, Svetkey, Suchindran, Willard and Ginsburg2014; Peck et al., Reference Peck, Borle, Folkersen and Austin2022).
Communication of genetic information in families in Mendelian genetics is essential to enable cascade testing for at-risk family members. Family communication has been extensively studied especially in monogenic settings (Burns et al., Reference Burns, James and Ingles2018; Gaff et al., Reference Gaff, Clarke, Atkinson, Sivell, Elwyn, Iredale, Thornton, Dundon, Shaw and Edwards2007; Mendes et al., Reference Mendes, Metcalfe, Paneque, Sousa, Clarke and Sequeiros2018). As PGS information does not have the same family implications as monogenic diseases, these issues are potentially less critical, but careful communication by health professionals regarding the implications for family members is needed where monogenic and polygenic information is combined to provide a refined personalized risk.
Ethical, Legal and Social Issues (ELSI) in PGS
Implementation of PGS in clinical care and population health will necessitate consideration of ethical, legal and social issues (ELSI) against a background of existing international ELSI literature, policy and regulation regarding genetics, and genomics more broadly. A key consideration is whether PGS raise new or unique ELSI considerations. It has been proposed that PGS give rise to similar ELSI issues as occurs in other kinds of genetic and genomic testing, although in a modified and distinct way (A. C. F. Lewis & Green, Reference Lewis and Green2021). Examples of PGS ELSI issues include equity, using PGS in reproduction, and possible insurance discrimination.
Equity considerations in the use of PGS comprise two aspects: access to testing and the impact of the use of PGS on existing social determinants of health. Any widespread clinical or population health implementation of PGS must take place alongside effective efforts to ensure that this application will benefit recipients regardless of ancestral background or other socio-demographic factors. It is essential that existing inequities in access to monogenic testing are not further exacerbated. Current GWAS and genomic databases do not represent the diversity of human genomes, leading to inequities due to lack of PGS availability or reduced predictive ability (Martin et al., Reference Martin, Kanai, Kamatani, Okada, Neale and Daly2019). In addition, many of the common, complex conditions where PGS have been reported to predict risk are impacted to a large degree by social determinants of health, such as living conditions and income. Resources should not be diverted to PGS as a technological solution to entrenched health problems while failing to address these well-described and longstanding issues. Instead, strategies to implement PGS should include active consideration of how this transition can contribute to the ongoing work of improving health services and social structures for the unde-served and marginalized.
The HGSA does not endorse the use of PGS in reproductive decision making, including preconception screening, embryo testing and prenatal diagnosis (K. W. Davis, Reference Davis2021; Forzano et al., Reference Forzano, Antonova, Clarke, de Wert, Hentze, Jamshidi, Moreau, Perola, Prokopenko, Read, Reymond, Stefansdottir, van El and Genuardi2022). Not only do current PGS provide information on the risk of a fraction of likely future medical disorders, and in many cases a modest component of the heritability for those disorders, but many of the conditions where PGS might provide a prediction of future risk can be effectively mitigated or prevented through health behavior modification. Using PGS to select against the risk of conditions (including common adult-onset disease) is currently beyond the scope of sustainable reproductive care. Irrespective of whether the use of PGS data is eventually shown to have an element of clinical utility in the reproductive setting, any use of this information should be informed by the values of those seeking this information, including the rationale for use of PGS and what reproductive decisions might be made in light of results obtained (Forzano et al., Reference Forzano, Antonova, Clarke, de Wert, Hentze, Jamshidi, Moreau, Perola, Prokopenko, Read, Reymond, Stefansdottir, van El and Genuardi2022).
Insurance providers are aware of PGS and seem enthusiastic about its potential future use (including as a ‘leading’ source of information) in risk assessments for products such as life insurance and income-protection insurance (Vukcevic & Chen, Reference Vukcevic and Chen2018; Scott McKay, Reference Scott McKay2022). The HGSA’s current position statement on insurance does not explicitly consider the use of PGS in research and clinical practice (Newson et al., Reference Newson, Ayres, Boyle, Gabbett and Nisselle2018). However, the HGSA recognises the need for education on ELSI aspects of insurance, and recommends that the Australian government take a more active role in regulating use of genetic information in personal insurance, which can include PGS. It is now the HGSA’s recommendation that the Australasian insurance industry deliberate and transparently disclose how information derived from PGS will be used, especially because risk rating for personal insurance products has always incorporated the notion of polygenic inheritance. PGS should also be considered in future revisions of any relevant Industry policy, such as the existing Australian Industry led moratorium (Financial Services Council, 2019). In a separate position statement, the HGSA promotes liaison between regulators, the insurance industry genetics profession to foster accurate interpretation and use of genetic information, especially for emerging test types such as PGS. The Society also advocates that genetic information obtained in research studies is excluded and that government should play a more active role in regulating the use of genetic information in insurance (HGSA, 2023)
Additional ELSI considerations relate to the technology itself. While array-based approaches that provide hundreds of thousands of genotypes from a single test are cost effective, their output can potentially be used to simultaneously generate PGS and define risk for many different conditions. As with any genetic test, testing should have a well-justified indication and a mode of consent appropriate to the test circumstance should be sought. Consent to generate PGS should include engagement over the purpose of the test and what information it might generate. The possibility of PGS generating unanticipated information should also be discussed as part of any strategy to implement PGS testing in practice. While genetic tests used for PGS can be designed to mitigate the identification of unexpected information, the need to incorporate information about an individual’s ancestry to correctly interpret a PGS means that laboratories may test for ancestry (explicitly or implicitly) as part of the PGS calculation. Issues around measuring and interpreting genetic ancestry are complex (A. C. F. Lewis et al., Reference Lewis, Molina, Appelbaum, Dauda, Di Rienzo, Fuentes, Fullerton, Garrison, Ghosh, Hammonds, Jones, Kenny, Kraft, Lee, Mauro, Novembre, Panofsky, Sohail, Neale and Allen2022) and laboratories and clinicians need to consider the implications of generating, reporting and retaining genetic-ancestry data; for instance, if the tested ancestry differs from the reported ancestry, would this be disclosed, and if so, how could this be done in a way consistent with patient-centred practice?
To the HGSA’s knowledge, PGS have not been specifically mentioned in relevant regulation to date, such as in pathology accreditation guidelines. We encourage relevant bodies to consider PGS when revising such instruments. As with other genomic technologies, care should be taken to ensure that commercial imperatives or drivers to increased information provision are not the determining factors in the implementation of PGS if this is not genuinely reflected in the clinical or personal utility of that information.
Behavioral Response to PGS Testing
Generally in public health even effective behavior change interventions typically have modest effects with significant heterogeneity of short- and long-term outcomes (R. Davis et al., Reference Davis, Campbell, Hildon, Hobbs and Michie2015). For diseases such as cancer and cardiovascular disease, genetic testing for monogenic causes has been shown to lead to increased risk-mitigating behaviors (Heshka et al., Reference Heshka, Palleschi, Howley, Wilson and Wells2008). Early studies that investigated similar responses for genomic testing were limited by methodological issues (Hollands et al., Reference Hollands, French, Griffin, Prevost, Sutton, King and Marteau2016) and found only equivocal evidence that PGS-based tests resulted in behavioral changes associated with prevention or early detection of disease (Frieser et al., Reference Frieser, Wilson and Vrieze2018; Hollands et al., Reference Hollands, French, Griffin, Prevost, Sutton, King and Marteau2016).
Several studies have now evaluated the impact of communicating PGS on health behavior, with mixed results (Wallingford et al., Reference Wallingford, Kovilpillai, Jacobs, Turbitt, Primiero, Young, Brockman, Soyer, McInerney-Leo and Yanes2022). Compared to individuals with a low PGS, those with a high PGS have reported improvements in sun protection behaviors and skin examinations (Lacson et al., Reference Lacson, Doyle, Qian, Del Rio, Forgas, Valavanis, Carvajal, Gonzalez-Calderon, Kim, Roetzheim, Sutton, Vadaparampil and Kanetsky2021; Saya et al., Reference Saya, McIntosh, Winship, Clendenning, Milton, Oberoi, Dowty, Buchanan, Jenkins and Emery2020), and increased uptake of risk-reducing medication for cardiovascular disease, resulting in lowered LDL-cholesterol levels (Muse et al., Reference Muse, Chen, Liu, Fernandez, Schrader, Molparia, León, Lee, Pubbi, Mejia, Ren, El-Kalliny, Prado Montes de Oca, Aguilar, Ghoshal, Dias, Evans, Chen, Zhang and Torkamani2022). However, other studies in type 2 diabetes and cardiovascular disease have shown mixed results with regard to physical activity, weight loss and smoking cessation (Godino et al., Reference Godino, van Sluijs, Marteau, Sutton, Sharp and Griffin2016; Widen et al., Reference Widen, Junna, Ruotsalainen, Surakka, Mars, Ripatti, Partanen, Aro, Mustonen, Tuomi, Palotie, Salomaa, Kaprio, Partanen, Hotakainen, Pöllänen and Ripatti2022). The extent to which PGS-based tests lead to positive behavioral responses requires further investigation as an important determinant of their value in clinical and public health settings. To date, there has been limited use of health behavior theory to inform PGS intervention, with most studies focusing on education as a key driver of behavior change (Wallingford et al., Reference Wallingford, Kovilpillai, Jacobs, Turbitt, Primiero, Young, Brockman, Soyer, McInerney-Leo and Yanes2022). While important, education alone is not sufficient to drive behavior change. Thus, careful consideration of health behavior theory will be required to identify barriers and facilitators of behavior change and develop targeted interventions based on PGS.
Education and Communication Needs for PGS Implementation
The clinical application of PGS is wide ranging, extending the impact of genomics to many more common disorders beyond the scope of established Mendelian genetics. There is the potential for substantially more clinicians to become involved in ordering, interpreting and explaining PGS information. Although this will involve introducing novel concepts and information to health professionals that have had limited exposure to genetics, healthcare professionals are used to managing technically and conceptually complex medical information; hence it is important to avoid the notion of genetic exceptionalism — the concept that genetic information is entirely unique and fundamentally different from other kinds of medical information (Garrison et al., Reference Garrison, Brothers, Goldenberg and Lynch2019; Mannette, Reference Mannette2021).
Workforce implications require consideration, including the extent of the role that the relatively small workforce of clinical geneticists and genetic counsellors will be able to play. The breadth of applications of PGS indicate that education and communication will need to move beyond the domain of specialist genetic healt care professionals. Models have been proposed in which general practitioners would have a primary role in the provision of PGS, with support from genetic health professionals (A. C. F. Lewis & Green, Reference Lewis and Green2021). However, even experienced health professionals currently involved in familial cancer risk assessment have reported low levels of confidence and knowledge around interpreting and communicating PGS (Smit et al., Reference Smit, Sharman, Espinoza, Wallingford, Young, Dunlop, Tiller, Newson, Meiser, Cust and Yanes2021), pointing to a widespread need for further education.
The development of practice guidelines, risk prediction tools, decision support and online point-of-care risk communication tools and resources will be key in supporting PGS integration into clinical practice and population health (Smit et al., Reference Smit, Newson, Keogh, Best, Dunlop, Vuong, Kirk, Butow, Trevena and Cust2019; Wallingford et al., Reference Wallingford, Kovilpillai, Jacobs, Turbitt, Primiero, Young, Brockman, Soyer, McInerney-Leo and Yanes2022). Evidence from international settings where PGS-based tests are more available have found reluctance to utilize these tests due to common concerns about the absence of such clinical guidelines, as well as insufficient evidence of clinical utility and inequity for patients from non-European backgrounds (McGuinness et al., Reference McGuinness, Fassi, Wang, Hacking and Ellis2021). This suggests the need for a broad scope of education that includes not just the technical aspects of the interpretation of PGS but a focus more generally on the benefits and limitations of this type of testing (Slunecka et al., Reference Slunecka, van der Zee, Beck, Johnson, Finnicum, Pool, Hottenga, de Geus and Ehli2021; Torkamani et al., Reference Torkamani, Wineinger and Topol2018).
Equally, efforts to improve public understanding of PGS are also required. An extensive literature exists around effective communication of genetic risk information for monogenic conditions. This evidence suggests that genetic counseling, lifestyle counseling, and written patient information can also improve patient understanding of polygenic risk information (Fenton et al., Reference Fenton, Smit, Keogh and Cust2018; Kaur et al., Reference Kaur, Meiser, Yanes, Young, Barlow-Stewart, Roscioli, Smith and James2019; Wallingford et al., Reference Wallingford, Kovilpillai, Jacobs, Turbitt, Primiero, Young, Brockman, Soyer, McInerney-Leo and Yanes2022; Yanes, Kaur, et al., Reference Yanes, Kaur, Meiser, Scheepers-Joynt, McInerny, Barlow-Stewart, Antill, Salmon, Smyth, James and Young2020). Patient understanding is also linked to a clinician’s own familiarity with the field, with evidence for significantly higher comprehension in patients who had PGS explained by a trained health professional (Haga et al., Reference Haga, Barry, Mills, Svetkey, Suchindran, Willard and Ginsburg2014). In many applications PGS will be combined with information about other risk factors, adding to the complexity of counseling by requiring appropriate contextualization of different risk factors, their contribution and potential for modification. Scalable models are required that balance patient communication needs and preferences with effective health service delivery (Wallingford et al., Reference Wallingford, Kovilpillai, Jacobs, Turbitt, Primiero, Young, Brockman, Soyer, McInerney-Leo and Yanes2022). Triaged approaches have been proposed that vary the level of PGS information and risk management recommendations, depending on an individual’s level of risk (Smit et al., Reference Smit, Reyes-Marcelino, Keogh, Dunlop, Newson and Cust2020). Early studies have found individualised approaches, such as face-to-face communication for high-risk results, and letters or emails for low-risk results, are preferred by patients (Ghanouni et al., Reference Ghanouni, Sanderson, Pashayan, Renzi, von Wagner and Waller2020).
Summary and Future Directions
There is broad consensus that PGS have the potential to be useful and impactful in clinical practice and public health, although further data are required on several fronts to demonstrate clinical utility. Clinical utility is a subjective and summative assessment and must be established within each context of use, considering disease, population, stakeholder and system-specific determinants (Moorthie, Reference Moorthie, Hall, Janus, Brigden, Babb de Villers, Blackburn, Johnson and Kroese2021). Research focused on clinical utility should be complemented by the development and evaluation of necessary infrastructure, including regulatory frameworks, standardized methodology, validation and reporting protocols, and multilevel education and communication initiatives (see Figure 1). For many conditions, sufficient preclinical data exist to warrant commencement of implementation research to better understand how PGS will function within various care pathways and to ensure that future clinical implementation occurs in a timely, equitable, and ethical manner supported by the high standards of evidence expected in clinical care.

Fig. 1. Potential strategies to enable PGS to be implemented in clinical practice.
Conclusion
PGS hold great promise for use within healthcare and are currently being examined in multiple clinical settings as well as in research. There is already a significant body of evidence from Mendelian genetics that can inform PGS implementation, preventing duplication and resource wastage. At the current time, however, PGS are not ready for widespread implementation into clinical practice or population health.
Box 2. HGSA Recommendations for PGS
Recommendations
Recognizing the potential of PGS to lead to new knowledge and improved health outcomes the HGSA currently:
-
1. Supports further research to explore the potential benefits and harms of specific applications of PGS, to identify applications that can effectively and equitably improve human health, and to design appropriate implementation strategies that engage consumers, health professionals, and policy makers to ensure that education, ethical, legal and social issues are all addressed.
-
2. Acknowledges that PGS are currently being accessed in clinical care and recommends clinicians ensure that they understand the limitations of current PGS detailed above, proceeding with caution.
-
3. Strongly supports the inclusion of more diverse populations and collaboration between multiple disciplines in future PGS research
-
4. Recognizes that implementation of PGS will require the dedication of resources to workforce planning and education, and the development of clinical aids such as practice guidelines and point of care decision support tools.
-
5. Does not support the use of PGS in preimplantation genetic testing and/or prenatal diagnosis currently due to significant scientific, health system, and ELSI concerns.
Acknowledgments
We thank the members of the Education, Ethics, and Social Issues Committee and other members of the Human Genetics Society of Australasia who reviewed the statement. This statement was reviewed and approved by the HGSA Council in January 2023. We also thank Ms Jane Tiller for her advice about insurance during the drafting of this position statement.
Data availability statement
N/A.
Financial support
TY is supported by an NHMRC EL1 Investigator Grant # 2009136. AEC is supported by a NHMRC Investigator Grant #2008454
Conflict of interest
None.
Ethical standards
N/A.






