INTRODUCTION
Noroviruses are recognized as the most common cause of viral gastroenteritis outbreaks [Reference Lopman1]. The clinical symptoms, including vomiting, diarrhoea, nausea and abdominal pain, last for 12–60 h [Reference Kaplan2]. The incubation period is 24–48 h [Reference Kaplan2]. The main transmission route is faecal–oral, although person-to-person, airborne and fomite transmissions have been reported [Reference Parashar3]. Since October 2003, Sweden has had nationwide voluntary laboratory-based norovirus surveillance. From 2004 to 2008 the annual number of microbiologically confirmed norovirus cases ranged from 1447 to 6651 (S. Rubinova, personal communication). A Swedish study showed that 101 food- and waterborne norovirus outbreaks resulted in more than 4100 cases between January 2002 and December 2006 [Reference Lysen4].
On 14 November 2007, the Department of Epidemiology, Swedish Institute for Infectious Disease Control, was notified by the Department of Communicable Disease Control and Prevention, Stockholm County, of gastroenteritis in employees of a manufacturing company. All ill employees were working in the research and development unit, which consisted of about 2000 office workers. Exploratory interviews showed that cases had visited different food outlets in the affected unit 1 or 2 days prior to onset of illness. Furthermore, no cases from other units of the company or from the community were reported. Therefore, we hypothesized that the exposure took place on 12 or 13 November either via a food outlet or water. The hypotheses were tested by conducting epidemiological, microbiological and environmental investigations. The objective of the investigations was to identify the source and agent of infection in order to prevent possible future outbreaks.
METHODS
Epidemiological investigation
The investigation consisted of two retrospective cohort studies, where the results of the first study led to the second study. The first study was conducted among all office workers from the affected unit to test for an association between a particular food outlet and illness. The second study was conducted among all office workers from the affected unit who visited the company canteen to test for an association between specific food items eaten and illness.
A link to a standardized web-based questionnaire was sent, via the Department of Human Resources, to all employees in the affected unit. Data were collected on clinical symptoms, exposure to different food outlets (i.e. canteen, kiosk, café, food vending machine) and water consumption. Following the results of the first questionnaire, a link to a second questionnaire was sent to those who reported visiting the canteen and provided a valid email address. This questionnaire listed specific food items and beverages.
The descriptive analysis was conducted using a sensitive case definition, where a case was defined as a person working in the affected unit with diarrhoea, vomiting, nausea or abdominal pain after 18:00 hours (12 November). For both cohorts the analytical study was conducted using a specific case definition. Using a specific case definition we attempted to exclude probable secondary cases [Reference Makary5]. A case was more specifically defined as a person working in the affected unit with onset of diarrhoea and/or vomiting between 11:00 hours (13 November) and 13:30 hours (14 November).
Data were analysed using Stata version 10.0 (StataCorp LP, USA). Using univariate analysis risk ratios (RR) with 95% confidence intervals (CI) were calculated for different food outlets and specific food items and beverages. A P value <0·05 was considered significant. Respondents who answered ‘don't know’ or did not reply at all to an exposure were excluded from the analysis of that particular exposure. Multivariable binary regression analysis was performed to evaluate potential interaction and to control for confounding. Only risk factors with a univariate P value <0·2 were tested in multivariable analysis.
Microbiological and environmental investigation
Stool samples from food handlers and office workers were collected. The samples were tested for viruses at the Centre for Microbiological Preparedness, Swedish Institute for Infectious Disease Control. Initial tests for norovirus genogroups I and II were performed using reverse transcription single-round multiplex PCR with the primers described by Yan et al. [Reference Yan6]. Subsequent tests for norovirus genogroup I were performed using the same PCR, but with the primers published by Gallimore et al. [Reference Gallimore7]. Sequence analysis was performed for nororvirus genotyping. The capsid fragment was sequenced using the BigDye Terminator cycle-sequencing kit (Applied Biosystems) and run on an automated sequencer (Applied Biosystems ABI Prism 3100 DNA sequencer). The sequences were edited by the Seqman II module in the Lasergene software package (DNAStar Inc., USA). Genotyping was done using the Quick genotyping database of the Foodborne Viruses in Europe (FBVE) network [Reference Duizer8]. The samples were also tested for other viruses (adenovirus, sapovirus, rotavirus), routinely tested bacteria (Salmonella, Campylobacter, Yersinia, Shigella), toxin-producing bacteria (enterotoxigenic Escherichia coli, Staphylococcus aureus, Clostridium perfringens) and parasites (Giardia, Cryptosporidium, Entamoeba histolytica).
An environmental assessment of the kitchen of the company canteen was performed. This assessment included sampling of tap water and food items. The tap water was tested for indicator bacteria (coliforms, Escherichia coli). The food items were tested for indicator bacteria (Escherichia coli, Enterobacteriaceae, Enterococcus) and toxin-producing bacteria (Staphylococcus aureus, Clostridium perfringens, Bacillus cereus). Culture methods were used for these bacteriological investigations. The tap water and food samples were not tested for viruses.
RESULTS
Epidemiological investigation
Concerning the descriptive study, a link to the questionnaire was sent to 2003 office workers of whom 1744 (87%) responded. Of 1744 respondents 413 (attack rate 24%) met the sensitive case definition and 306 (attack rate 18%) met the specific case definition. Of 413 cases the onset of symptoms ranged from 20:30 hours (12 November) to 06:00 hours (17 November) (Fig. 1). The peak of the outbreak was between 18:00 and 24:00 hours (13 November). Eighty-seven per cent of the cases were male. The median age was 36 years (range 22–65 years). The attack rate in men was 25% and in women 18% (P=0·02). Reported symptoms were nausea (92%), fatigue (90%), vomiting (80%), diarrhoea (77%), abdominal pain (72%), headache (65%), fever (64%), and myalgia (47%). The mean duration of illness was 21 h (range 2–120 h).

Fig. 1. Date and time of symptom onset in cases during a norovirus outbreak, Sweden, 2007. ![]() , Cases with diarrhoea and/or vomiting between 11:00 hours (13 November) and 13:30 hours (14 November) (n=306).
, Cases with diarrhoea and/or vomiting between 11:00 hours (13 November) and 13:30 hours (14 November) (n=306). ![]() , Cases with diarrhoea, vomiting, nausea or abdominal pain after 18:00 hours (12 November) (n=99).
, Cases with diarrhoea, vomiting, nausea or abdominal pain after 18:00 hours (12 November) (n=99).
In the first analytical study, univariate analysis demonstrated that the highest RR of 27·1 (95% CI 15·7–46·8) was for office workers visiting the company canteen on 12 November. The attack rate in visitors was 37·5% (288/767) compared to 1·4% (13/938) in non-visitors. Of all cases 94% reported visiting the company canteen.
In the second analytical study, 730 office workers who reported visiting the company canteen on 12 November were sent a link to the questionnaire listing specific food items and beverages served that day. There were 615 (84%) respondents of which 236 (attack rate 38%) met the specific case definition. In univariate analysis, several food items were significantly associated with illness (Table 1). The highest RR of 3·6 was for consumption of tomatoes. Of all cases 83% reported eating tomatoes from the salad buffet. Interaction between tomato and hamburger consumption was significant (P<0·001). In a model including tomato consumption, hamburger consumption and a term for interaction between these two food items, the RRs for tomato and hamburger consumption were 5·6 (95% CI 3·2–9·6) and 4·9 (95% CI 2·4–9·8), respectively. The interaction term was 0·2 (95% CI 0·1–0·5). There was no significant interaction between tomato consumption and any of the other food items with a univariate P value <0·2. Furthermore, none of these food items remained significantly associated with illness after adjusting for tomato consumption.
Table 1. Univariate analysis of risk of illness for employees by different food items served in a company canteen on 12 November 2007, Sweden
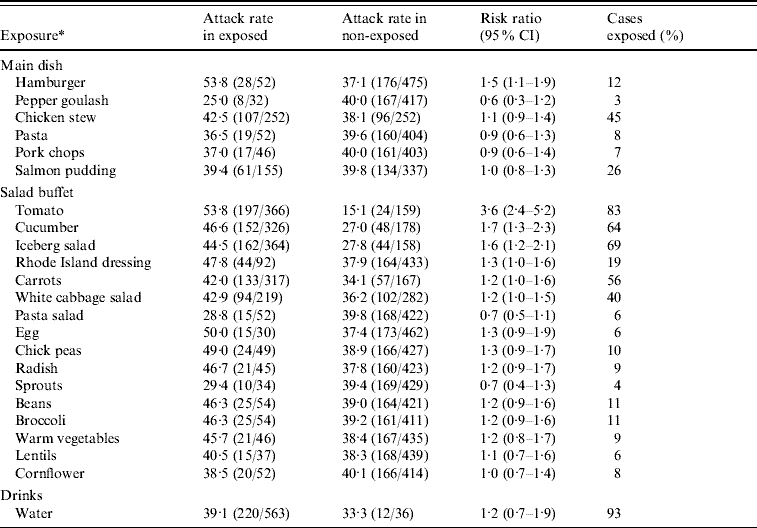
CI, Confidence interval.
* Excluded those food items with less than 20 persons exposed: fillet of beef, carrot soup, sushi, Italian dressing, pepsi, zingo, apple juice, lingon drink, beer.
Microbiological and environmental investigation
Sixteen of 17 food handlers provided a stool sample of which two reported gastroenteritis. One of these food handlers reported having vomited at the workplace on 12 November. This food handler worked that morning until 10:00 hours and cut the tomatoes for the salad buffet and prepared the ingredients for the hamburger buffet (i.e. slicing bread, cutting tomatoes) before vomiting in the lavatory. The lunch started at 11:00 hours after the food handler had vomited and gone home. Norovirus genogroup I was identified in the sample from this food handler. Further characterization of the norovirus strain demonstrated genotype GI.3 (Desert Shield). Another food handler reported having had gastroenteritis on 8 November. The sample from this food handler tested negative for bacteria and no material was left to test for norovirus. Six of 16 samples were tested for viruses using the initial method. With this method all six samples were negative. Another six samples were tested using the subsequent method. With this method four samples were positive for norovirus genogroup I. It was possible to further characterize two of these samples as GI.3. Three of four positive samples were from food handlers who reported not having had gastroenteritis, and one was from the food handler who vomited. The samples which tested positive arrived at the virology laboratory on 21 November. The food handler who vomited was asymptomatic at the time of sampling. Nine of 16 samples were routinely tested for bacteria and one sample was tested for toxin-producing bacteria. All bacteriological tests produced a negative result. One of 16 samples was tested for parasites and was also negative.
Twelve office workers who reported gastroenteritis provided a stool sample. Nine of 12 samples were tested for viruses using the initial method. Using this method all nine samples were negative. Repeated testing of three samples using the subsequent method showed that all three samples were positive for norovirus GI.3. These samples arrived at the virology laboratory on 22 November. Three of 12 samples were routinely tested for bacteria and one sample was tested for toxin-producing bacteria. All bacteriological tests produced a negative result. One of 12 samples was tested for parasites and was also negative.
The environmental assessment of the kitchen revealed that the number of sinks for hand washing was insufficient. The tap water and the following food items, which were saved from the lunches on 12 and 13 November, were sampled on 15 November: chicken casserole, ham casserole, lamb casserole, spare ribs, boiled rice, salmon pudding and alfalfa sprouts. All samples tested negative for bacteria.
DISCUSSION
Over 400 office workers from the same unit of a company fell ill during a foodborne outbreak caused by norovirus GI.3. Canteen visitors on 12 November had an increased risk of illness compared to non-visitors. An investigation of canteen visitors' consumption of particular food items showed that tomatoes from the salad buffet and hamburgers were the most likely vehicles of infection. Norovirus GI.3 was identified in stool samples from three office workers and from a food handler who vomited after cutting the tomatoes for the salad buffet and handling the hamburger ingredients.
The hamburgers were served in a buffet with freshly cut tomatoes as one of the ingredients to choose from. Freshly cut tomatoes were also served in a separate salad buffet. Tomatoes may logically be suspected to be the only vehicle as hamburgers are often eaten with tomatoes. Nevertheless, another explanation might be that another hamburger ingredient was contaminated, as the food handler also cut the bread for the hamburgers, and not restricted solely to the tomatoes. Therefore, canteen visitors who did not eat their hamburger with tomatoes could still have been infected. We did not obtain information regarding whether or not the canteen visitors had chosen tomatoes as their hamburger ingredient. Therefore, we can not rule out the other hamburger ingredient as possible vehicle of infection.
The analytical study shows that tomatoes from the salad buffet and hamburgers were associated with illness. None of these food items remained from the lunch on 12 November for sampling. Although other food items from the lunches on 12 and 13 November were sampled, it was not possible to test for norovirus because of the unavailability of an assay. However, methods for detection of norovirus in foods are improving and norovirus has been detected in oysters and raspberries [Reference Nenonen9, Reference Le Guyader10].
The strain identified in this outbreak belonged to norovirus genotype GI.3. Although this strain has previously been associated with foodborne outbreaks [Reference Johansson11, Reference Sala12], norovirus genogroup I strains do not seem to be as frequently associated with outbreaks as norovirus genogroup II strains [Reference Glass13]. The virological analysis of the faecal samples initially failed to identify norovirus. It could be that norovirus could not be detected because of a mismatch in the PCR primers for genogroup I. This was a problem during the previous Swedish norovirus genogroup I outbreak [Reference Johansson11] and shows that it is necessary to follow-up negative stool samples with additional primer sets [Reference Vinje14]. Another explanation could be that the subsequent method is more sensitive and can detect norovirus in samples with a lower viral load. The food handler who vomited was asymptomatic at the time of sampling. Although norovirus can be detected up to 3 weeks after the symptomatic phase, the virus is then excreted in lower amounts [Reference Rockx15]. Timely sampling might have facilitated the microbiological investigation.
The microbiological investigation demonstrated that faecal samples from a symptomatic food handler and three office workers had identical norovirus sequences corresponding to GI.3. Three other food handlers who reported not having gastroenteritis also tested positive for norovirus genogroup I. The identification of norovirus in both food handler and cases has been reported before [Reference Vivancos16]. It is unknown whether the food handler who reported having had gastroenteritis on 8 November was positive for norovirus and back at work on 9 November. If so, this food handler might have transmitted the virus to the other food handlers during the post-symptomatic phase. Of the four food handlers who tested positive, one reported vomiting after preparing the tomatoes for the salad buffet and hamburger ingredients. It could be that these food items were contaminated in the pre-symptomatic phase.
Food handlers with vomiting and/or diarrhoea should to be excluded from work and not return until 48 h after recovery [Reference Schmid17, Reference LeBaron18]. The food handler went home after vomiting and vomited after preparing the food items. It is unknown if the food handler experienced any nausea while handling the food items. Norovirus excretion in pre-symptomatic food handlers and in food handlers with mild symptoms has been reported previously [Reference Vivancos16, Reference Gaulin19]. In these reports, the food handlers reported vomiting and/or diarrhoea after preparing and serving the food items. In this outbreak, the food handler vomited after preparing, but before serving the food items. The outbreak could have been prevented if the food items prepared by the food handler some hours before vomiting had not been served.
ACKNOWLEDGEMENTS
We thank Harry Vennema, Dutch National Institute for Public Health and the Environment (RIVM), for guidance in the identification of norovirus. We thank Viviane Bremer (EPIET), Alicia Barrasa Blanco (EPIET), Francisco Luquero (Epicentre) and Inga Velicko (SMI) for critical feedback. The EPIET fellowship of T. P. Zomer was funded by the European Commission DG SANCO.
DECLARATION OF INTEREST
None.




