Introduction
The comparison of DNA sequence data, enabled especially by the availability of sequences in large public sequence databases, can help us to resolve phylogenetic relationships and reveal cryptic taxa, contributing to a broader understanding of evolutionary and ecological patterns and processes (e.g. Ito et al. Reference Ito, Robledo, Iharlegui and Tanaka2016, Webster et al. Reference Webster, Emami-Khoyi, van Dyk, Teske and Jansen van Vuuren2020). The unique flora of the remote and isolated sub-Antarctic islands are fairly well characterized (e.g. Green & Walton Reference Greene and Walton1975, Lord Reference Lord2015, Chau et al. Reference Chau, Mtsi, Münbergová, Greve, le Roux and Mairal2020, Siljak-Yakovlev et al. Reference Siljak-Yakovlev, Lamy, Takvorian, Valentin, Gouesbet, Hennion and Robert2020). However, although some molecular phylogenetic studies have been conducted (e.g. Wagstaff et al. Reference Wagstaff, Breitwieser and Ito2011, Lehnebach et al. Reference Lehnebach, Winkworth, Becker, Lockhart and Hennion2017), the evolutionary origins of many of the constituent species remain obscure.
Marion Island (46.9°S, 37.75°E) is one of two volcanic islands in the Prince Edward Island archipelago in the southern Indian Ocean, ~1770 km south-east of South Africa. Low herbaceous vegetation covers much of the island below the high unvegetated peaks, growing in the cool, wet conditions that characterize the sub-Antarctic terrestrial region (Smith & Mucina Reference Smith, Mucina, Mucina and Rutherford2006). Due to high precipitation, aquatic habitats, including mires, streams, lakes and rock pools, are common throughout the lowlands of the island (Hänel & Chown Reference Hänel and Chown1998). Although several species of mosses and vascular plants grow on saturated substrates or are emergent or floating-leaved in standing water, only two species listed in the vascular plant flora grow submersed on the bottom of waterbodies, namely Limosella australis R.Br. (Scrophulariaceae) and Ranunculus moseleyi Hook.f. (Ranunculaceae) (Gremmen & Smith Reference Gremmen, Smith, Chown and Froneman2008). Limosella australis was first recorded on Marion Island by botanist Brian Huntley during the first extensive floristic survey of the island on the 1965–1966 South African Biological and Geological Expedition to Marion and Prince Edward Islands (Huntley Reference Huntley, Van Zinderen Bakker, Winterbottom and Dyer1971) and has been collected sporadically since then. Ranunculus moseleyi was also first recorded on Marion Island during the 1965–1966 Expedition (Huntley Reference Huntley, Van Zinderen Bakker, Winterbottom and Dyer1971), but only one herbarium specimen of this species from Marion Island is known to exist (B.J. Huntley 1568, AAS) and its current occurrence there has been put in doubt (Lehnebach et al. Reference Lehnebach, Winkworth, Becker, Lockhart and Hennion2017, Chau et al. Reference Chau, Mtsi, Münbergová, Greve, le Roux and Mairal2020).
During the South African National Antarctic Programme relief voyage to Marion Island in April 2018, two morphologically distinct populations of submersed aquatic plants were observed, one growing in small rock pools and the other in the nearshore of a large lake. Specimens examined from the populations were past flower or sterile, but the vegetative characters of the rock pool population conformed to the description for L. australis, with small linear leaves (Cook Reference Cook2004). The other population from the lake resembled the description of R. moseleyi, with longer leaves differentiated into petiole and blade (Hooker Reference Hooker1879). In this study, we aimed to clarify the taxonomic identities of these two forms of submersed aquatic plants on Marion Island through genetic sequencing of nuclear and plastid loci and comparison of sequences with those in public sequence databases.
Materials and methods
Sampling
Specimens from two populations of submersed aquatic vascular plants were collected on Marion Island in April 2018 (Fig. 1a & Table I). Leaf tissues were preserved on silica gel and voucher specimens were deposited at the University of Pretoria herbarium (PRU). The two populations differed in vegetative morphology and habitat. In one population, small plants with linear leaves < 25 mm long grew submersed in a small, shallow rock pool on grey lava (J.H. Chau 331; Fig. 1b). In the other population, larger plants with leaves that were linear or had a long petiole and small spatulate blade (up to 80 mm long in total) grew submersed on the muddy nearshore of the large lake Prinsloo Meer (J.H. Chau 333; Fig. 1c). We also sampled a herbarium specimen determined as L. australis with linear leaves, some with a slightly spatulate tip, collected from a small pool on black lava on Marion Island (W.A. Haddad 1023; Fig. 1d).

Fig. 1. Distribution and morphology of sampled Limosella specimens from Marion Island. a. Collection localities on Marion Island. b. J.H. Chau 331 c. J.H. Chau 333 d. W.A. Haddad 1023.
Table I. Collection and voucher information for sampled specimens from Marion Island and GenBank accession numbers for sequences. ITS = internal transcribed spacer; PRU = University of Pretoria herbarium.

Molecular methods and analyses
Total genomic DNA was extracted from dried leaf tissue using a modified cetyltrimethylammonium bromide (CTAB) protocol (Doyle & Doyle Reference Doyle and Doyle1987). Three loci were chosen for sequencing based on the availability of sequences from Limosella and Ranunculus in public sequence repositories and their ability to resolve interspecific relationships in these groups (Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017, Lehnebach et al. Reference Lehnebach, Winkworth, Becker, Lockhart and Hennion2017). The nuclear ribosomal internal transcribed spacer (ITS) region was amplified using the primers AB101 and AB102 (Robinson et al. Reference Robinson, Harris and Juniper2001). The plastid trnL-trnF intergenic region and rps16 intron were amplified with the primers c and f (Taberlet et al. Reference Taberlet, Gielly, Pautou and Bouvet1991) or rpsF and rpsR2 (Oxelman et al. Reference Oxelman, Lidén and Berglund1997), respectively. Polymerase chain reaction (PCR) amplification was conducted in a thermocycler using the PCR programme in Zietsman et al. (Reference Zietsman, Dreyer and Jansen van Vuuren2009) and an annealing temperature of 50–52°C. PCR amplicons were cleaned before use as templates in cycle sequencing reactions with the same primers used in PCR to sequence both forward and reverse directions. Cycle sequencing products were read on an ABI 3730 Genetic Analyzer at the Stellenbosch University Central Analytical Facilities DNA Sequencing Unit (Stellenbosch, South Africa).
Sequence chromatograms were checked for quality and assembled into contigs in Geneious v9.1.6 (Biomatters), from which consensus sequences were extracted. Each consensus sequence was queried in a Basic Local Alignment Search Tool (BLAST) search against the National Center for Biotechnology Information (NCBI) GenBank nucleotide collection database (accessed 9 October 2019). Sequences of the top 100 BLAST search hits as sorted by bit score were retrieved. Additional sequences of the three loci from Limosella in the NCBI GenBank nucleotide database were identified by searching the terms ‘Limosella’ and ‘ITS’, ‘trnL’ or ‘rps16’, and any sequences that were missing from the BLAST search results were retrieved. These sequences, together with the sequences from the Marion Island specimens, were aligned using MAFFT v7 with default parameters (Katoh & Standley Reference Katoh and Standley2013). Phylogenetic trees were inferred with each sequence matrix under the maximum likelihood optimality criterion using RAxML v8.2 (Stamatakis Reference Stamatakis2014) under the generalized time-reversible (GTR) + gamma model of nucleotide substitution with support values obtained from 100 rapid bootstrap replicates. Trees were rooted according to phylogenetic relationships inferred for Scrophulariaceae in Oxelman et al. (Reference Oxelman, Kornhall, Olmstead and Bremer2005). We also inferred a phylogenetic tree from a concatenated plastid dataset comprising all Marion Island samples and samples of Limosella aquatica, Limosella australis, Limosella curdieana and Limosella major having sequences of both trnL-trnF and rps16 using the analysis parameters above. As an additional line of evidence, for each locus, species-specific polymorphisms (i.e. sites that are variable between species but fixed within species) were identified and compared with the sequences from Marion Island specimens.
Results
The nuclear ITS locus was successfully sequenced in all three sampled Marion Island specimens, with the total length of the consensus sequence ranging from 516 to 751 bp. We also sequenced the plastid locus trnL-trnF in all three specimens with lengths ranging from 915 to 944 bp, and rps16 in two specimens (J.H. Chau 331 could not be amplified) with lengths ranging from 877 to 886 bp. For all loci, sequences from all three Marion Island specimens were identical where they overlapped, except for trnL-trnF, in which the specimen W.A. Haddad 1023 had a duplication of 16 bp in length. Sequences are deposited in GenBank (Table I).
For all three loci, the most similar BLAST search hits in the NCBI GenBank database to the Marion Island specimens were from Limosella, with the remaining top hits being from other genera in the family Scrophulariaceae. Phylogenetic analyses placed the Marion Island specimens within a clade comprising only and all Limosella accessions with very strong support (bootstrap percentage (BP) = 100%) for all three loci (Figs 2 & S1–S3). More narrowly, for ITS, the Marion Island specimens were in a strongly supported clade (BP = 93%) with accessions of L. aquatica L., L. curdieana F.Muell. and L. major Diels (Figs 2a & S1). For trnL-trnF, relationships within the Limosella clade in general had weak support, largely as a result of relatively few characters supporting branching patterns (i.e. short branch lengths). The Marion Island specimens were in a slightly narrower well-supported clade with accessions of L. africana Glück, L. aquatica, L. australis, L. curdieana, L. grandiflora Benth., L. macrantha R.E.Fr. and L. major, but a poorly supported clade comprising L. aquatica, L. curdieana, L. macrantha and L. major excluded the Marion Island specimens (Figs 2b & S2). For rps16, the Marion Island specimens were in a strongly supported clade (BP = 81%) with accessions of L. australis (Figs 2c & S3). The concatenated plastid tree similarly placed the Marion Island specimens in a strongly supported clade (BP = 100%) with L. australis (Fig. S4).
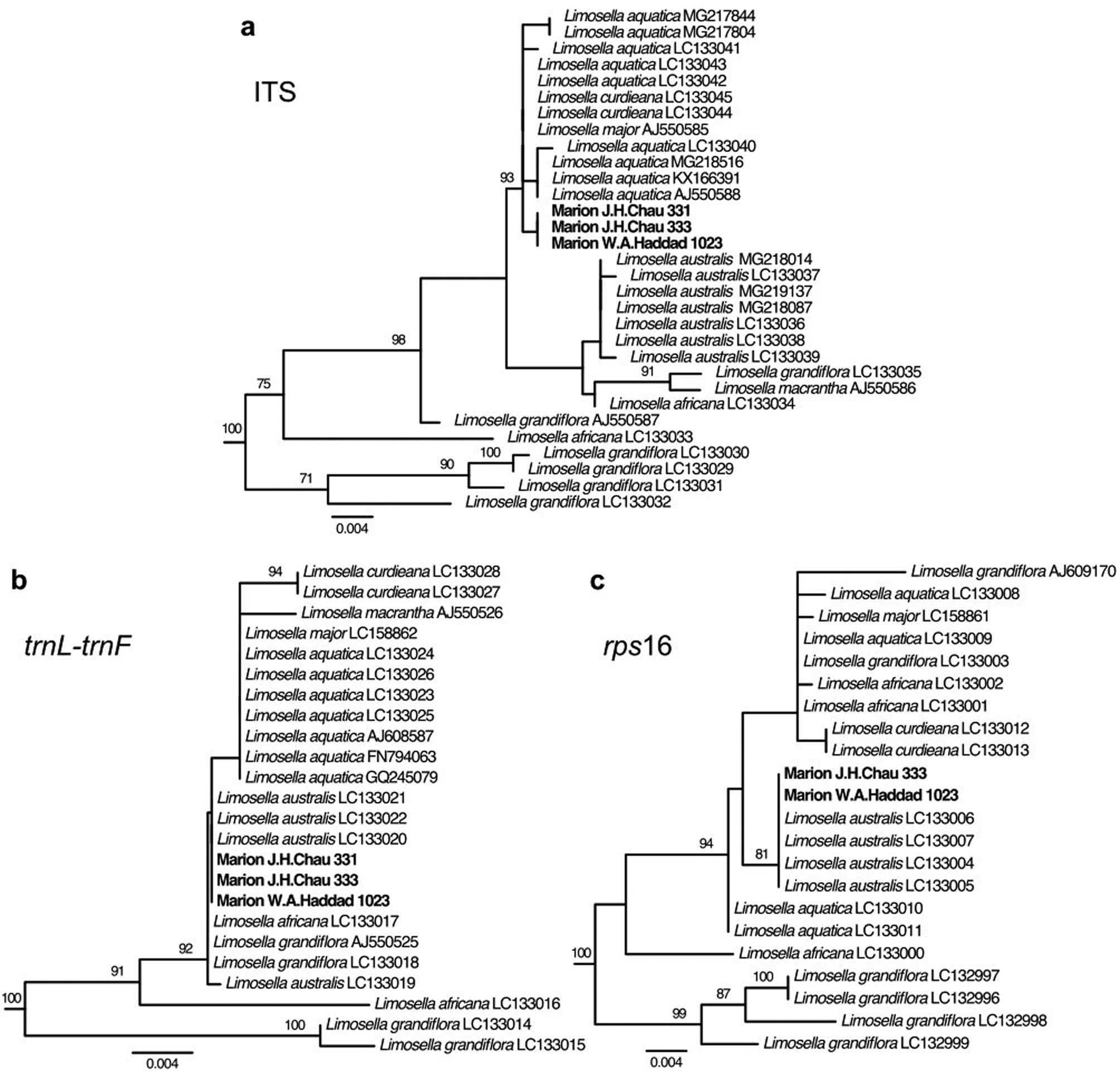
Fig. 2. Phylogenetic trees from maximum likelihood analyses of DNA sequence data from Marion Island specimens and from the National Center for Biotechnology Information (NCBI) GenBank database. Taxa outside Limosella have been pruned from trees. Species and GenBank accession numbers are shown at the tips. Bootstrap support values for nodes > 70% are shown on the branches. a. Nuclear internal transcribed spacer (ITS); b. plastid trnL-trnF; c. plastid rps16.
Based on our phylogenetic results, we focused our examination of species-specific sequence polymorphisms on the clade comprising the species L. aquatica, L. curdieana and L. major (hereafter termed the L. aquatica clade) vs L. australis. For ITS, we retrieved from the GenBank database nine sequences for L. aquatica, two sequences for L. curdieana, one sequence for L. major and seven sequences for L. australis (Table SI). There were seven single-nucleotide polymorphisms (SNPs) and three insertions/deletions (indels) that were fixed within the L. aquatica clade and L. australis, but differed between them. The Marion Island sequences matched that of the L. aquatica clade at all of these variant sites and additionally had one unique SNP that differentiated the Marion Island sequences from both the L. aquatica clade and L. australis (Table II). For trnL-trnF, we retrieved seven sequences for L. aquatica, two sequences for L. curdieana, one sequence for L. major and four sequences for L. australis (Table SI). One SNP and one indel were fixed within the L. aquatica clade and L. australis, but differed between them. The Marion Island sequences matched that of L. australis at these two variant sites (Table III). For rps16, we retrieved four sequences for L. aquatica, two sequences for L. curdieana, one sequence for L. major and four sequences for L. australis (Table SI). There were two indels that were fixed within the L. aquatica clade and L. australis, but differed between them. The Marion Island sequences matched that of L. australis at these two positions. In addition, there was one indel variant that was fixed within L. aquatica only, with a different allele found in L. australis, L. curdieana and L. major. In this case, the Marion Island sequences matched those of L. australis, L. curdieana and L. major (Table IV).
Table II. Species-specific sequence polymorphisms in the Limosella aquatica clade (L. aquatica, L. curdieana, L. major), L. australis and Marion Island specimens in the nuclear internal transcribed spacer (ITS) locus, listed by their position in the sequence alignment. The number of sequences examined for each species or clade is given in parentheses.

Table III. Species-specific sequence polymorphisms in the Limosella aquatica clade (L. aquatica, L. curdieana, L. major), L. australis and Marion Island specimens in the plastid trnL-trnF locus, listed by their position in the sequence alignment. The number of sequences examined for each species or clade is given in parentheses.

Table IV. Species-specific sequence polymorphisms in the Limosella aquatica clade (L. aquatica, L. curdieana, L. major), L. australis and Marion Island specimens in the plastid rps16 locus, listed by their position in the sequence alignment. The number of sequences examined for each species or clade is given in parentheses.

a At this position, L. curdieana (two sequences) and L. major (one sequence) share the same sequence variant as L. australis instead of L. aquatica.
Discussion
Identification of specimens from Marion Island
Genetic evidence unequivocally identifies all of our specimens (J.H. Chau 331, J.H. Chau 333, W.A. Haddad 1023) of submersed aquatic plants from Marion Island as members of the genus Limosella. The specimens are nearly identical genetically, despite differing vegetative morphologies. Loci from the plastid genome (trnL-trnF, rps16) place the Marion Island specimens as members of L. australis, which is the only species of Limosella currently recognized in the flora of Marion Island (Gremmen & Smith Reference Gremmen, Smith, Chown and Froneman2008, Chau et al. Reference Chau, Mtsi, Münbergová, Greve, le Roux and Mairal2020). However, a nuclear locus (ITS) places the specimens in a clade with L. aquatica, L. curdieana and L. major. As the chloroplast is typically contributed only by the female gamete, this suggests that the maternal lineage of the Marion Island specimens is L. australis, while ancestry from the L. aquatica clade is also present.
Evidence of cytonuclear discordance suggests that Limosella on Marion Island has a history of hybridization or introgression. Discrepancy in the phylogenetic position of specimens between plastid and nuclear gene trees has been interpreted as evidence of hybridization in other plants, including aquatic groups (Tippery & Les Reference Tippery and Les2011, Ito et al. Reference Ito, Robledo, Iharlegui and Tanaka2016). Other possible causes of gene tree incongruence include incomplete lineage sorting and paralogous genes (Maddison Reference Maddison1997). However, the fact that a clear pattern of incongruence is found only in the Marion Island specimens suggests that it reflects an evolutionary event specific to the Marion Island population, rather than more species-wide phenomena such as the retention of ancestral polymorphisms or gene duplication. The other L. australis sequences from public sequence databases included in our analyses come from specimens from New Zealand, the Falkland Islands/Malvinas, Ecuador and Canada, while the sequences of L. aquatica are from North American, European and Asian specimens, the sequences of L. curdieana come from Australian specimens and the sequences of L. major are from African specimens (Table SI). In no other sequences from these widely dispersed specimens did we see the same pattern of gene tree incongruence reported here for the Marion Island specimens.
The vegetative morphology of the Marion Island specimens shows characteristics of both putative parental species, L. australis and a member of the L. aquatica clade. The leaves of L. australis are linear, without a distinct blade, cylindrical and typically up to 30 mm long and 2 mm wide (Cook Reference Cook2004, Crow & Hellquist Reference Crow and Hellquist2005, Barringer Reference Barringer2019). The leaves of L. aquatica are flat, with a distinct petiole (up to 100 mm) and a lanceolate to spatulate blade (up to 30 mm long and 30 mm wide) (Crow & Hellquist Reference Crow and Hellquist2005, Barringer Reference Barringer2019). The leaves of L. curdieana and L. major are similarly differentiated into petiole and blade (Cook Reference Cook2004, Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017). The different specimens of Limosella on Marion Island encompass these two forms, with J.H. Chau 331 fitting the description of L. australis with short, linear leaves, J.H. Chau 333 fitting the description of L. aquatica with longer leaves with a small spatulate blade and W.A. Haddad 1023 being intermediate with linear leaves, some with a slightly broader tip. The different forms of Limosella on Marion Island may be due to phenotypic plasticity. The specimens with linear leaves were found growing in small rock pools, whereas the larger specimen with leaves with a differentiated blade was from the nearshore of Prinsloo Meer, a large coastal lake adjacent to a king penguin colony. The waters of Prinsloo Meer are likely to be more nutrient-rich compared to those in rock pools, allowing plants in the former to grow larger leaves with greater differentiation of the blade and petiole. In addition, the habitats varied in water depth, which may affect the development of leaf size and shape. In the greater depths of Prinsloo Meer, plants are consistently submersed and may have access to less light compared to the plants in rock pools, which are very shallow and may occasionally leave plants only partially submersed. It is also possible that genetic variability in a hybrid population results in different genetically determined vegetative morphology, although we detected almost no genetic variation among specimens in the three loci we sequenced. Another possible explanation for the different morphological forms is that they represent different developmental stages. In L. aquatica × L. subulata E.Ives (= L. australis) hybrids reported from Great Britain, the leaves were intermediate in shape compared to the parental species, showed considerable variation and displayed developmental changes in morphology, being ‘successively subulate and lanceolate-spathulate with a long petiole’ (Vachell & Blackburn Reference Vachell and Blackburn1939). The age of our specimens from Marion Island are unknown; however, the smaller specimen with linear leaves (J.H. Chau 331) appeared to have fruits and thus to be reproductively mature. Experimental studies in common gardens and a more thorough survey of Limosella on Marion Island would illuminate the causes of morphological variation in the population.
The ploidy level of L. australis has been reported as diploid (2n = 20), whereas L. aquatica is tetraploid (2n = 40) (Vachell & Blackburn Reference Vachell and Blackburn1939, Les Reference Les2018). In L. aquatica × L. australis hybrids reported from Great Britain, the intermediate triploid (2n = 30) condition was seen (Vachell & Blackburn Reference Vachell and Blackburn1939). Cytogenetic studies of the plants on Marion Island should be conducted to further elucidate the putative history of hybridization or introgression in the population.
Evolutionary implications of hybridization
The timing of hybridization with respect to the arrival of Limosella on Marion Island is unknown. We found no specimens on Marion Island that are purely L. australis or L. aquatica with all loci unanimously aligning with one species, although more extensive sampling on the island should be conducted to confirm their absence. This makes it unlikely that hybridization is occurring presently on Marion Island. However, it is possible that both species arrived on Marion Island in the past and formed hybrids before populations of the parental species declined. Limosella australis is currently distributed in several other sub-Antarctic islands, South America, eastern North America, Great Britain, Australia, New Zealand and possibly southern Africa (Cook Reference Cook2004, Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017, Barringer Reference Barringer2019, https://vicflora.rbg.vic.gov.au). The distribution of L. aquatica spans northern South America, North America, northern Africa, Europe and Asia (Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017, Barringer Reference Barringer2019), while L. curdieana is found in Australia and New Zealand (https://vicflora.rbg.vic.gov.au) and L. major is in eastern and southern Africa (Cook Reference Cook2004, Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017). Areas of overlap between the range of L. australis and the ranges of L. aquatica, L. curdieana and L. major, where hybrids may have formed and whence dispersed to the sub-Antarctic, include parts of South America and North America, Great Britain, Australia, New Zealand and possibly southern Africa. In fact, a population of L. aquatica × L. australis hybrids has been identified in Wales, Great Britain, based on intermediate morphology and karyotype (Vachell & Blackburn Reference Vachell and Blackburn1939).
Hybridization has been proposed as a mechanism for adaptation to novel and marginal habitats and the evolution of traits such as invasiveness (Ellstrand & Schierenbeck Reference Ellstrand and Schierenbeck2000, Kawecki Reference Kawecki2008). In L. aquatica × L. australis hybrids from Great Britain, greater vegetative vigour, more rapid growth and better winter survival were observed compared to the parental species, though the hybrids were sterile (Vachell & Blackburn Reference Vachell and Blackburn1939, Stace Reference Stace and Stace1975). The genetic variation and novelty created by hybridization may have contributed to the ability of the Limosella population on Marion Island to establish and grow, but this hypothesis should be further tested with common garden experiments with plants from Marion Island and species from other areas. If hybrids are sterile, Limosella is still able to reproduce clonally by producing runners and stolons (Cook Reference Cook2004), through which heterosis in hybrid genotypes could be maintained and propagated (Ellstrand & Schierenbeck Reference Ellstrand and Schierenbeck2000). Although a specimen of Limosella was found with what appeared to be fruits (J.H. Chau 331), no seeds were observed and it is unknown whether any viable gametes were produced. Asexual reproduction and high vagility of vegetative propagules, which are especially common in aquatic plants, may allow for greater persistence and spread of hybrid individuals in these plants (Les & Philbrick Reference Les and Philbrick1993). Although there are few records of hybrid plants in the sub-Antarctic (e.g. Acaena decumbens × tenera on South Georgia, Walton & Greene Reference Walton and Greene1971), more focused surveys and studies on the prevalence of hybrids in the sub-Antarctic flora, as well as other groups of organisms, would shed light on the importance of this evolutionary phenomenon in such an extreme environment.
Taxonomic and biogeographical perspectives
Morphological variation and phylogenetic incongruence in the Limosella specimens from Marion Island add to the need for a contemporary revision of the taxonomy of the genus in the sub-Antarctic and globally. The species L. australis, including its application to sub-Antarctic specimens, had previously been synonymized as a variety of L. aquatica (var. tenuifolia Hook.f.) partly because leaf shape, a character used for differentiating the two species, was thought to be too variable, with the range of leaf shapes displayed in L. aquatica encompassing the subulate form of L. australis (Hooker Reference Hooker1847, Hearn Reference Hearn1901). Later botanists disagreed, recognizing discontinuities in the morphological range, and reinstated L. australis as a distinct species (Pearsall Reference Pearsall1934). The specimens on Marion Island demonstrate that within apparently genetically homogeneous populations, morphological variability can exist in traits often used in species identification. Cytonuclear discordance has also been detected in another species of Limosella, L. macrantha, from tropical Africa, in which plastid DNA placed it with L. major, while nuclear ITS resolved it as sister to an accession of L. africana (Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017). As a whole, the genus Limosella was last comprehensively treated in the early twentieth century (Glück Reference Glück1934). A recent molecular phylogenetic study including 8 of 12–15 recognized species clarified some relationships (Ito et al. Reference Ito, Tanaka, Albach, Barfod, Oxelman and Muasya2017), but with unresolved ambiguity in the geographical distribution, morphological range and taxonomic circumscription of species, a global revision of the group is necessary (Cook Reference Cook2004, Smithies Reference Smithies, Germishuizen, Meyer, Steenkamp and Keith2006, Les Reference Les2018, Barringer Reference Barringer2019).
Limosella australis is also recorded in the floras of other Southern Ocean islands, including the Falklands/Malvinas, Prince Edward, Crozet, Kerguelen and Auckland islands (Lord Reference Lord2015). In a global study of the species, all examined specimens from the Southern Ocean, including from Crozet, Kerguelen and Auckland islands, were placed in L. australis, based on the morphology of the style, which was claimed to be the only character consistently separating the species from L. aquatica (Lourteig Reference Lourteig1964). Limosella specimens from Kerguelen were determined to have a ploidy of 6× (2n = 60), which is double the chromosome number of L. australis × L. aquatica hybrids from Great Britain (3×, 2n = 30), suggesting that this sub-Antarctic population may be an allopolyploid (Vachell & Blackburn Reference Vachell and Blackburn1939, Siljak-Yakovlev et al. Reference Siljak-Yakovlev, Lamy, Takvorian, Valentin, Gouesbet, Hennion and Robert2020). Additional genetic studies on plants from the other islands should be conducted in order to determine whether the hybrid lineage is endemic to Marion Island or found in other parts of the sub-Antarctic and to resolve the relationship of the Marion Island population with other populations, which would shed light on patterns of biogeographical connections in the sub-Antarctic region.
Status of Ranunculus moseleyi on Marion Island
None of our specimens of submersed aquatic plants were identified as Ranunculus based on genetic data, even though the vegetative morphology of some specimens (J.H. Chau 333) fits the species description of R. moseleyi: ‘narrow, linear leaves, sometimes spatulate at the ends’ (Gremmen & Smith Reference Gremmen and Smith2004). However, this morphology could also match that of L. aquatica or possibly even L. australis. It has been noted that in the flora of the Prince Edward Islands and Kerguelen Island, R. moseleyi and Limosella are very easily confused if flowers or fruits are not present (Gremmen & Smith Reference Gremmen and Smith2004). The absence of R. moseleyi among our specimens adds weight to the lack of current evidence for the presence of the species on Marion Island. We know of only one herbarium specimen of R. moseleyi from Marion Island, from the 1965–1966 South African Biological and Geological Expedition to Marion Island (B.J. Huntley 991, AAS). The presence of aggregates of follicular fruits confirms this determination. The only other specimen we found that had a determination of R. moseleyi on the label (B.J. Huntley 2048, PRE), also from the 1965–1966 Expedition, later had its determination changed to L. australis, which is consistent with the presence of capsular fruits, as in Limosella but not Ranunculus, on the specimen. Smith & Mucina (Reference Smith, Mucina, Mucina and Rutherford2006) noted that the last record of R. moseleyi on Marion Island was in 1987, but we are not aware of any other vouchers for which taxonomic identification can be confirmed. In more recent floristic surveys on the island, R. moseleyi has not been found (Raath-Krüger et al. Reference Raath-Krüger, McGeoch, Schöb, Greve and le Roux2019, P. le Roux, personal communication 2020), and the species was left off a recent plant list for Marion Island (Chau et al. Reference Chau, Mtsi, Münbergová, Greve, le Roux and Mairal2020).
Supplementary material
One supplemental table and four supplemental figures can be found at https://doi.org/10.1017/S0954102021000079
Acknowledgements
We acknowledge the South African National Antarctic Program (SANAP) and the Department of Environmental Affairs for logistical support on the relief voyage to Marion Island. Permission to collect was granted by the Department of Environmental Affairs (permit number 1/2013). We thank the herbaria at the University of Pretoria (PRU) and the South African National Biodiversity Institute (PRE) for access to specimens, and we thank Peter le Roux and Elsa van Ginkel for helpful discussions about the plants on Marion Island. We appreciate helpful comments from the editor and two reviewers that improved our manuscript.
Author contributions
JHC and MG did the fieldwork and sampling. JHC performed the laboratory work and data analyses and wrote the manuscript with support and assistance from MG and BJvV.
Financial support
We gratefully acknowledge financial support from the National Research Foundation (SANAP grants 110728 and 110734).
Details of data deposit
DNA sequences are available on GenBank under the accession numbers in Table I.









