The utilisation and availability of proteins depend on the types of proteins and their specific susceptibility to enzymatic hydrolysis in the gastrointestines, and are also highly associated with protein molecular structures and profiles(Reference Yu1, Reference Doiron, Yu and McKinnon2). Protein secondary structures include mainly α-helix and β-sheet, and small amount of β-turn and random coil(Reference Kemp3–Reference Marinkovic and Chance6). The protein molecular structure profiles (such as mid-IR molecular absorption intensities and ratios of these secondary structures) and molecular spectral characteristics of protein structure amide I and amide II and their ratio may influence protein quality, nutrient utilisation, availability and digestive behaviour in both animals and human subjects(Reference Wetzel, Srivarin and Finney7–Reference Yu10), mainly because protein structure affects access to microbial degradation and gastrointestinal digestive enzymes, which affects protein values and protein availability (total intestinally absorbed protein supply).
However, studies on protein structures at molecular and cellular levels in relation to nutrient availability and digestive behaviours of proteins are still limited compared with traditional animal nutrition research. The protein molecular structure profile could be significantly affected by heat (autoclaving) processing(Reference Doiron, Yu and McKinnon2) and gene transformation(Reference Yu, Jonker and Gruber11).
Recently, the different types of new co-products from bioethanol processing, wheat dried distillers grains with solubles (wheat DDGS), triticale DDGS, pea DDGS, hull-less barley DDGS, maize DDGS and blend DDGS (e.g. wheat:maize = 70:30; 50:50; 20:80), were produced in North America(Reference Nuez-Ortín and Yu12). The variation in nutrient profiles between and within DDGS types is attributed to grain type, grain varieties, processing methods and/or processing conditions. During bioethanol processing, ethanol fermentation removes most of the starch from grain kernels and concentrates the other components, mainly protein, fibre, fat and minerals in DDGS. Additionally, the DDGS are heat dried, which affects the protein molecular structure. So far, none of the published studies in the literature reported how the molecular structure changes in the DDGS are associated with nutrient availability in the rumen and intestine in dairy cattle. None of the published studies in the literature reported the possibility of using the protein molecular structure profile as a predictor of the nutrient value of the co-products. The DDGS can be a major source of ruminally undegraded protein (RUP) in diets, but it can vary greatly(Reference Nuez-Ortín and Yu12). The protein molecular structure profiles might account for some of this large variation and would improve the estimation of protein value of different types of DDGS.
The objectives of this novel study were to investigate the relationship between protein molecular structures (in terms of protein α-helix and β-sheet intensity and their ratio and amide I to amide II intensity and their ratio) and protein rumen degradation kinetics (rate and extent), estimated protein intestinal digestibility, degraded protein balance(Reference Tamminga, Van Straalen and Subnel13) and total truly absorbed protein in small intestine (metabolisable protein)(Reference Tamminga, Van Straalen and Subnel13) in different types of DDGS. The hypothesis of this preliminary study was that the differences in protein molecular structures in bioethanol co-products were associated with the various measures of protein degradability and digestibility, which could be used as predictors of protein nutrient availability.
Materials and methods
The experiments were carried out at the Saskatchewan Structure Sciences Center and Livestock Research Station at the University of Saskatchewan.
The animal experiment (protocol no. 19910012) was reviewed and approved by the Animal Care Committee of the University of Saskatchewan, and was conducted in accordance with the guidelines of the Canadian Council on Animal Care(14).
Co-products of bioethanol production, original cereal grains and nutrient analysis
The different types of bioethanol co-products, wheat DDGS, maize DDGS and blend DDGS (wheat:maize = 70:30), and original feedstock wheat and maize samples from bioethanol plants were used for the present study. The detailed sampling structure and collection time frames were described previously in the publication by Nuez-Ortín & Yu(Reference Nuez-Ortín and Yu12) in 2009. The detailed methods and calculations for chemical and nutrient analyses(15–19), Cornell net carbohydrate and protein system (CNCPS) protein fractionation(Reference Sniffen, O'Connor and Van Soest20, Reference Chalupa and Sniffen21), in situ rumen degradation(Reference Ørskov and McDonald22–Reference Dhanoa24), estimated intestinal protein digestion(Reference Calsamiglia and Stern25) and modelling total nutrient supply(Reference Yu, Goelema and Leury26, Reference Yu, Christensen and McKinnon27), in dairy cattle fed the bioethanol co-products were reported by Nuez-Ortín & Yu in 2010(Reference Nuez-Ortín and Yu28, Reference Yu, Niu and Damiran29). In the present study, the chemical, nutrient and structure profile data of the six different types of DDGS and original grain samples (summarised in Table 1)(Reference Nuez-Ortín and Yu12, Reference Nuez-Ortín and Yu28, Reference Yu, Niu and Damiran29) were used in a correlation study between protein molecular structure profiles (amide I, amide II, amide I to amide II ratio, α-helix, β-sheet and α-helix to β-sheet ratio) and nutrient availability.
Table 1 Summary of chemical, nutrient and protein molecular structure profiles of different grains (wheat, maize) and different types of dried distillers grains with solubles (DDGS) (wheat DDGS, maize DDGS and blend DDGS (wheat:maize=70:30) with total sample number n 6) from bioethanol production* (data sources (Reference Nuez-Ortín and Yu12, Reference Nuez-Ortín and Yu28, Reference Yu, Niu and Damiran29))

OM, organic matter; CHO, carbohydrate; NDF, neutral-detergent fibre; ADF, acid-detergent fibre; CP, crude protein; SCP, soluble crude protein; NPN, non-protein nitrogen; CNCPS, Cornell net carbohydrate and protein system; TDN, total digestible nutrient; NRC, National Research Council; NEL3X, net energy for lactation at production level of intake (3x); NEm, net energy for maintenance; NEg, net energy for growth; RUP, ruminally undegraded protein; dRUP, digestibility of RUP; DVE, truly digested protein in the small intestine; OEB, degraded protein balance; ENDP, endogenous CP losses; EE, ether extract; NDIP, neutral-detergent insoluble protein; ADL, acid-detergent lignin.
Range indicates the range in all data, not just treatment means.
* These data were used to study the relationship between protein molecular structures and protein availability.
† Carbohydrate was calculated as: carbohydrate = 100 − EE − CP − ash(19).
‡ Non-fibre carbohydrate = 100 − (NDF − NDIP) − EE − CP − ash(19).
§ Hemicellulose = NDF − ADF(19).
∥ Cellulose = ADF − ADL(19).
¶ Protein subfractions using CNCPS include PA = fraction of CP that is instantaneously solubilised at time zero; PB1 = fraction of CP that is soluble in borate-phosphate buffer and precipitated with TCA; PB2 = calculated as total CP minus the sum of fractions PA, PB1, PB3 and PC; PB3 = calculated as the difference between the portions of total CP recovered with NDF and ADF; PC = fraction of CP recovered with ADF, and is considered to be undegradable. It contained proteins associated with lignin and tannins and heat-damaged proteins such as the Maillard reaction products.
** True protein = PB1 (% of CP)+PB2 (% of CP)+PB3 (% of CP).
Diffuse reflectance IR Fourier transform spectroscopy
The DDGS samples were finely ground two times to pass through a 250 μm screen (Retsch ZM-1, Brinkmann Instruments (Canada) Limited, ON, Canada). Samples of the ground DDGS were then mixed with potassium bromide (IR grade; P5510, Sigma, St Louis, MO, USA) in a ratio of four parts co-product to one part potassium bromide in a 2 ml centrifuge tube and mixed by vortexing for several minutes. Diffuse reflectance IR Fourier transform spectroscopy was performed using a Bio-Rad FTS-40 with a ceramic IR source and mercury-cadmium-telluride (MCT) detector (Bio-Rad laboratories, Hercules, CA, USA). Data were collected using Win-IR software (Bio-Rad Digilab, Cambridge, MA, USA). Spectra were generated from the mid-IR (4000–800 per cm) portion of the electromagnetic spectrum with 256 co-added scans and a spectral resolution of 4 per cm(Reference Doiron, Yu and McKinnon2).
Molecular spectral analysis of protein amide I and amide II ratio and α-helix and β-sheet ratio
Molecular spectral analysis was done with OMNIC 7.2 software (Spectra Tech, Madison, WI, USA). Chemical functional groups in the grains and DDGS (protein amide I and amide II and protein secondary structures α-helix and β-sheet) were identified according to the published reports(Reference Marinkovic and Chance6, Reference Wetzel, Srivarin and Finney7, Reference Himmelsbach, Khalili and Akin30–Reference Budevska, Chalmers and Griffiths32). The protein IR spectrum has two primary features. The protein amide I bond is primarily C = O stretching vibration (80 %) plus C–N stretching vibration(Reference Jackson, Mantsch, Mantsch and Chapman5, Reference Jackson, Mantsch and Meyers33). Protein amide I absorbs at approximately 1655 per cm. Protein amide II which absorbs at approximately 1550 per cm consists primarily of N–H bending vibrations (60 %) coupled with C–N stretching vibrations (40 %)(Reference Jackson, Mantsch, Mantsch and Chapman5, Reference Jackson, Mantsch and Meyers33). The absorption intensity of peak area of amide I and amide II and their ratio were calculated with baseline region at approximately 1720–1485 per cm. The vibrational frequency of the protein amide I band is particularly sensitive to protein secondary structure(Reference Kemp3, Reference Marinkovic and Chance6, Reference Wetzel, Srivarin and Finney7, Reference Martin34, Reference Miller35). Protein α-helix is typically in the range of approximately 1648–1660 per cm and β-sheet is in the range of approximately 1625–1640 per cm. To estimate the intensity of α-helix and β-sheet, two steps were applied. The first step used Fourier self-deconvolution (a method for resolving intrinsically overlapped bands) and the 2nd derivative function in OMNIC 7.2 to obtain the Fourier self-deconvolution and 2nd derivative spectrum in protein amide I region at approximately 1720–1575 per cm only to identify protein amide I component peak frequencies. The detailed concepts and algorithm of Fourier self-deconvolution were described by Kauppinen et al. (Reference Kauppinen, Moffatt and Mantsch36) and Griffiths & Pariente(Reference Griffths and Pariente37). The second step was done to quantify the intensity of peak height of α-helix and β-sheet and their ratio(Reference Doiron, Yu and McKinnon2).
Statistical analysis
Correlation analysis
The relationship between the changes in protein structure profiles (in terms of α-helix to β-sheet ratio and amide I to amide II ratio) and changes in chemical and nutrient profiles (in terms of chemical composition, protein fractions, in situ rumen degradation kinetics, estimated intestinal digestion and predicted nutrient supply to cattle) in the different types of DDGS samples (no original grain data) were analysed using the CORR procedure of Statistical Analysis Systems (SAS version 9.1.3, SAS Institute Inc., Cary, NC, USA) with the FISHER option, which offers confidence limits and P-values for Pearson's correlation coefficients based on Fisher's z transformation. The normality tests were performed using UNIVARIATE procedure of SAS with NORMAL and PLOT options.
Multiple regression analysis
In order to determine which protein molecular structure parameters (amide I to amide II ratio and α-helix to β-sheet ratio) in the different types of DDGS samples (no original grain data) play an important role in determining protein utilisation and availability to cattle, a multiple regression analysis was carried out using the ‘PROC REG’ procedure of SAS (version 9.1.3) with a model as follows:
Model: Y = amide I to amide II ratio (R_I-II)+α-helix to β-sheet ratio (R_α-β).
The model variable selection used a ‘STEPWISE’ option with variable selection criteria: ‘SLENTRY = 0·05, SLSTAY = 0·05’. All variables left in the final model are significant at the 0·05 level. The residual analysis was carried out to test the regression model assumptions using UNIVARIATE procedure of SAS with NORMAL and PLOT options.
For all statistical analyses, significance was declared at P < 0·05 and trends (tendency) at P < 0·10. Differences among the treatments were evaluated using a multiple comparison test following Fisher's protected least significant difference method.
Results and discussion
The traditional approach to study protein value of different types of co-products from bioethanol production is to focus on total protein composition or total available protein composition using conventional ‘wet’ chemical analysis as suggested by National Research Council (NRC)-2001, which is called ‘a chemical summary approach’. However, using the NRC-2001 summary approach to determine the protein value of bioethanol co-products is not accurate. A fatal shortcoming of the NRC-2001 summary is that it does not consider the intrinsic structure and molecular make-up of the different types of co-products. Traditional ‘wet’ chemical analysis can determine total protein composition, but fails to detect protein inherent structure and molecular make-up due to destruction of the bioethanol co-product protein structure during the processing for chemical analysis. In a previous research, it was found that two barley varieties showed the same chemical composition, but significantly different digestive behaviour in ruminants. The two co-products had similar levels of RUP, but different availability of RUP. The NRC-2001 summary approach cannot address these issues. Therefore, a new approach that is able to account for structural variation at molecular and cellular levels between the different types of co-products from bioethanol production should be developed. This novel approach should provide a new concept and methodology for true protein value research. Our new approach in this preliminary study was to look at the protein molecular structure (amide I, amide II, α-helix and β-sheet structure profiles) in the different types of bioethanol co-products in relation to true protein value.
Protein structure amides I and II profiles in relation to nutrient profiles in bioethanol co-products
The amide I and amide II structure profiles depend on the protein molecular structural make-up, and can be affected by cereal grain variety(Reference Yu38), autoclave-heated processing conditions(Reference Doiron, Yu and McKinnon2) and gene transformation(Reference Yu, Jonker and Gruber11). Bioethanol processing involves enzymatic fermentation and heat drying of the DDGS products, which affects molecular structural characteristics of protein amide I and amide II and their ratios(Reference Yu, Niu and Damiran29). The differences in the amide I to amide II profiles and their ratio may be associated with differences in nutrient value. However, no study has been reported. Table 2 shows the correlations of protein structure amide I, amide II and amide I to amide II ratio with nutrient profiles in the DDGS. There were significantly positive correlations between protein structure amide I and amide II ratio and soluble crude protein (CP) with R 0·99 (P < 0·01), and a tended positive correlation with neutral-detergent insoluble protein with R 0·93 (P = 0·07), but no correlation (P>0·10) with acid-detergent insoluble protein (Table 2). These results indicate that higher protein amide I and amide II ratio associated with higher neutral-detergent insoluble protein in the bioethanol co-products. For Cornell net carbohydrate and protein system (CNCPS) protein subfraction correlation, the results showed that the protein amide I and amide II ratio had strongly positive correlations with the protein PA fraction (P < 0·01) with R 0·99 and a negative correlation with the protein PB2 subfraction (P = 0·04) with R − 0·95, but that it had no correlation (P>0·10) with the PB1, PB3 and PC fractions. For total digestible CP, the results showed that the protein amide I and amide II ratio had strongly positive correlation with total digestible CP (R 0·95, P = 0·05). (For explanation of protein subfractions, see Table 1.)
Table 2 Correlation between protein structures (amide I to amide II and their ratio) and chemical profiles, Cornell net carbohydrate and protein system (CNCPS) protein fractions, in situ degradation kinetics and nutrient supply in the different types of co-products (wheat dried distillers grains with solubles (wheat DDGS), maize DDGS and blend DDGS with total sample number n 4) from bioethanol production (include just dependent variables that were significantly affected by the ratios)

CP, crude protein; NDIP, neutral-detergent insoluble protein; NRC, National Research Council; DVE, truly digested protein in the small intestine; OEB, degraded protein balance.
* For explanation of protein subfractions, see Table 1.
For the correlation of in situ degradation parameters, the results showed that the protein amide I and amide II ratio had a correlation with the soluble fraction (R 0·94, P = 0·06), potentially degradation fraction (R 0·99, P < 0·01), undegradable fraction (R − 0·99, P < 0·01) and RUP ( − 0·96, P = 0·03), but that it had no correlation with degradation rate (P>0·10). The result indicated that lower protein amide I and amide II ratio was associated with a higher RUP value in DDGS. The protein amide I and amide II ratio had no correlation with estimated intestinal digestibility of RUP in vitro (P>0·10). For predicted nutrient supply correlation, the results showed that the protein amide I and amide II ratio had no correlation (P>0·10) with microbial protein synthesis absorbable microbial CP, endogenous CP losses, absorbable ruminally undegraded CP and total intestinally absorbed protein supply, but that it had strongly positive correlations with the rumen degraded protein balance (R 0·97, P = 0·02).
In a previous study, Doiron et al. (Reference Doiron, Yu and McKinnon2) reported that heating the Vimy flaxseed changed the chemical profiles, which decreased the soluble CP upon heating with a concomitant increase in non-protein N, neutral-detergent insoluble nitrogen and acid-detergent insoluble nitrogen. They found that the protein subfractions with the greatest changes were PB1 (fraction PB1 is a rapidly degradable protein fraction in the rumen), which showed a dramatic reduction, and PB2 (fraction PB2 is fermented in the rumen at a lower rate than buffer-soluble fractions and some of the PB2 fraction escapes to the lower gut), which showed a dramatic increase, demonstrating a decrease in the overall protein degradability. Their in situ results showed a reduction in rumen-degradable protein, but the intestinal digestibility analysed by a three-step in vitro method showed no changes in RUP. Modelling results showed that the heating increased total intestinally absorbable protein (feed DVE value) and decreased the degraded protein balance (feed OEB value), but that there were no differences between the treatments. There was a linear effect of heating time on the DVE and cubic effect on the OEB value. However, in their study(Reference Doiron, Yu and McKinnon2), no correlation study was done between amide I and amide II profiles and nutrient availability in Vimy flaxseed autoclaved at various conditions. No comparison could be made with the present study on the co-products of bioethanol processing.
Protein secondary structure profiles in relation to nutrient profiles in bioethanol co-products
The amide I band vibrational frequency can be used to determine the secondary structure of proteins because it is particularly sensitive to protein secondary structure(Reference Wetzel, Srivarin and Finney7, Reference Jackson, Mantsch and Meyers33, Reference Martin34, Reference Carey39). For the protein α-helix structure, the amide I typically is in the range of approximately 1648–1660 per cm. For β-sheet, the peak is in the range of approximately 1620–1640 per cm(Reference Marinkovic and Chance6, Reference Martin34). The amide II can also be used to assess the protein conformation and protein molecular chemical make-up. However, it arises from complex vibrations involving multiple functional groups, and it is less useful for protein structure prediction than the protein amide I band(Reference Jackson, Mantsch and Meyers33). In the DDGS, the spectrum of protein amide I original band shows peak centres at approximately 1658 and 1628 per cm. This was confirmed from both the Fourier self-deconvolution spectrum of amide I and the 2nd derivative spectrum of amide I at the region of approximately 1720–1575 per cm.
The DDGS differed in protein secondary structure conformation in terms of the ratio of protein α-helix and β-sheet, indicating the differences in protein molecular structural make-up and features. Bioethanol processing may change the protein molecular structure of the DDGS compared with the original products. These structural differences may impact the DDGS protein utilisation and availability in the rumen and intestine in ruminants.
Some microbial CP (such as yeast) that are present in the DDGS might affect the protein structure profiles of the DDGS, although the amount of microbial CP in the products is relatively small. During heat drying process, heat also denatures yeast, rendering them resistant to rumen degradation. While some of the protein content in the solubles is heated yeast(Reference Belyea, Rausch and Tumbleson40, Reference Klopfenstein, Erickson and Bremer41), only 20 % of them are ruminally degradable(Reference Herol42). Additionally, the solubles contribute to the RUP and absorbable ruminally undegraded CP contents in DDGS by providing simple sugars that increase the susceptibility to the Maillard reaction during heat drying(Reference Nuez-Ortín and Yu28). Dorion et al. (Reference Doiron, Yu and McKinnon2) reported that using the synchrotron radiation-based Fourier transformed IR microspectroscopy, and heating at 120°C for 40 and 60 min (not for 20 min) increased the protein structure α-helix to β-sheet ratio of Vimy flaxseed. There were linear effects of heating time on the ratio.
Table 3 shows the correlations of protein structure α-helix, β-sheet and α-helix to β-sheet ratio with nutrient profiles in the DDGS with P < 0·10. There was a tended correlation between the protein structure α-helix to β-sheet ratio and acid-detergent insoluble CP with R 0·91 (P = 0·09) (Table 3), but there were no correlations (P>0·10) between the protein structure α-helix to β-sheet ratio and non-protein N, soluble CP and neutral-detergent insoluble protein (data not shown). This result indicates that higher protein α-helix to β-sheet ratio is associated with a higher acid-detergent insoluble CP value in the bioethanol co-products.
Table 3 Correlation between protein structure α-helix to β-sheet ratio and chemical profiles, Cornell net carbohydrate and protein system (CNCPS) protein fractions, in situ degradation kinetics and nutrient supply in the different types of co-products (wheat dried distillers grains with solubles (wheat DDGS), maize DDGS and blend DDGS with total sample number n 4) from bioethanol production (include just dependent variables that were significantly affected by the ratios)
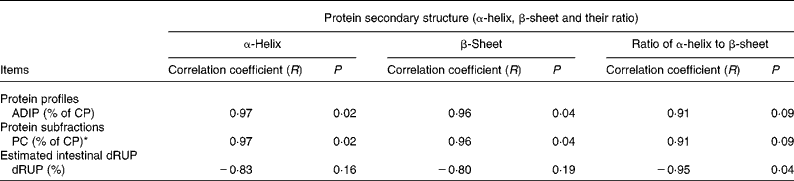
ADIP, acid-detergent insoluble protein; CP, crude protein; dRUP, digestibility of ruminally undegraded protein.
* PC = fraction of CP recovered with acid-detergent fibre, and is considered to be undegradable.
During bioethanol processing, heat drying process is applied. Heat drying facilitates the Maillard reaction, through which sugar residues condense with protein amino acids, rendering proteins indigestible. These indigestible proteins are recovered in the lignin and acid-detergent fibre. Thus, an indication of the severity of the drying conditions can be provided by the content of acid-detergent insoluble CP. A negative relationship between acid-detergent insoluble CP and the ruminal and intestinal availability of DDGS protein was reported(Reference Nuez-Ortín and Yu28). In the present study, a tended correlation between the α-helix to β-sheet ratio and acid-detergent insoluble CP was found, which may indicate that higher α-helix to β-sheet ratio may result in lower ruminal and intestinal protein availability of DDGS.
For Cornell net carbohydrate and protein system (CNCPS) protein fraction correlation, the results showed that the protein structure α-helix to β-sheet ratio had a positive correlation with the protein PC fraction (P = 0·09) with R 0·91, but that it had no correlation (P>0·10) with protein PA, PB1, PB2 and PB3 fractions. The PC fraction was undegradable, and contained proteins associated with lignin and tannins and heat-damaged proteins. These results indicate that a higher α-helix to β-sheet ratio may result in a higher undegradable protein content in the DDGS.
For in situ parameter correlation, the results showed that the protein α-helix to β-sheet ratio had no correlation with soluble fraction, degradable fraction, undegradable fraction, Kd (the rate of degradation of the D fraction; /h) and RUP. The results also showed that there was a strongly negative correlation between the protein α-helix to β-sheet ratio and estimated protein intestinal digestibility of RUP in vitro (R − 0·95, P = 0·04). These results suggest that a high protein α-helix to β-sheet ratio may be detrimental to the protein availability of the DDGS in the intestine in terms of its use as a feed ingredient. This result is opposite to the previous findings that indicate that a higher β-sheet content results in lower nutrient availability. However, the heating methods were different (autoclaving v. dry heat) and this might be a part of the reason.
For predicted nutrient supply correlation, the results showed that the protein α-helix to β-sheet ratio had no correlation (P>0·10) with absorbed microbial protein synthesis, endogenous CP losses, absorbable ruminally undegraded CP, total intestinally absorbed protein supply and degraded protein balance. However, in a previous study, the autoclaving of Vimy flaxseed changed the protein structure α-helix to β-sheet ratio, and decreased RUP and increased potential nutrient supply to dairy cattle. The protein structure α-helix to β-sheet ratio had a significantly positive correlation with total intestinally absorbed protein supply and a negative correlation with degraded protein balance(Reference Doiron, Yu and McKinnon2). These results suggest that different heating methods may have different impact on feed protein molecular structures.
Using protein structure profile as a predictor of the nutrient supply from bioethanol co-products
The results obtained from multiple regression analyses are shown in Table 4. The tested multiple regression model was: Y = ratio of α-helix to β-sheet+ratio of amide I to amide II. This analysis was done to find a suitable protein structure variable that can be used to predict nutrient supply from the DDGS to dairy cattle.
Table 4 Data obtained from regression analysis used to find the most important variables to predict protein nutrient supply using protein molecular structural parameters (α-helix to β-sheet ratio and amide I to amide II ratio) in the co-products (wheat dried distillers grains with solubles (wheat DDGS), maize DDGS and blend DDGS with total sample number n 4) from bioethanol production with tested regression model*
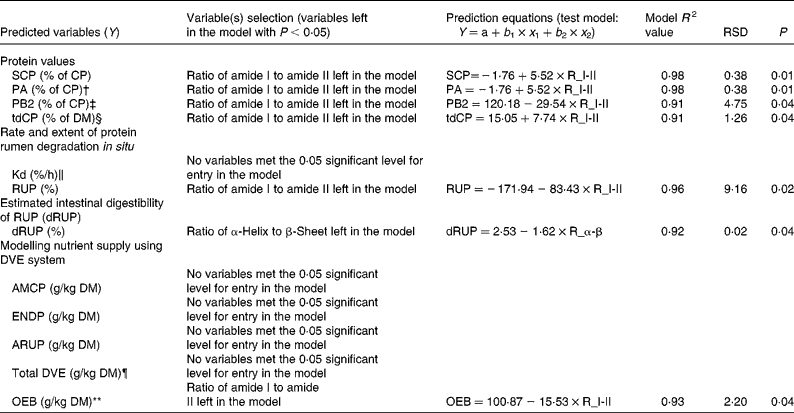
RSD, residual standard deviation; SCP, soluble crude protein; CP, crude protein; tdCP, total digestible CP; RUP, ruminally undegraded protein; dRUP, digestibility of RUP; AMCP, absorbable microbial CP; ENDP, endogenous CP losses; ARUP, absorbable ruminally bypassed CP; NRC, National Research Council; CNCPS, Cornell net carbohydrate and protein system.
* Model: Y = ratio of α-helix to β-sheet (R_α-β)+ratio of amide I to amide II (R_I-II).
† Protein subfractions using the CNCPS include PA (fraction of CP that is instantaneously solubilised at time zero).
‡ Protein subfractions using the CNCPS include PB2 (fraction of CP that is intermediately degraded in the rumen).
§ tdCP (NRC-2001 summary approach).
∥ Kd = in situ degradation rate.
¶ DVE = total intestinally absorbed protein supply.
** OEB = degraded protein balance.
The results showed that the ratio of α-helix to β-sheet was a better predictor of the estimated protein intestinal digestibility of the DDGS in vitro (with 92 % of the variance being accounted for). The ratio of amide I to amide II was a better predictor of the degraded protein balance of the DDGS (with 93 % of the variance being accounted for).
This is a novel approach to use protein molecular structure as a predictor of the protein nutrient availability in its preliminary stage. In order to obtain a more conclusive predictive equation, a large-scale in vivo study with various sources of bioethanol co-products is needed to test the applicability of the protein molecular structural parameters investigated. The development of a method to improve the estimation of protein value (protein degradability and digestibility) will highly benefit the scientific community.
Conclusions
In conclusion, the protein structure α-helix to β-sheet ratio in the DDGS had a strongly negative correlation with estimated intestinal digestibility of RUP, and it had no correlation with rumen degradation kinetics (rate and extent), total intestinally absorbed protein supply and degraded protein balance. The protein amide I to amide II ratio in the DDGS had a strongly positive correlation with the total digestible CP and strongly negative correlations with RUP and degraded protein balance, but it had no correlation with total intestinally absorbed protein supply.
The results indicate that the changes in the protein molecular structure α-helix to β-sheet ratio and the amide I to amide II ratio during bioethanol process, either due to enzymatic fermentation processing or due to final co-product drying, highly associated with estimated protein intestinal digestibility and degraded protein balance, can be used as predictor of the protein nutritive value in the DDGS sample from bioethanol processing. A large-scale in vivo study with various sources of bioethanol co-products is needed to test and verify the applicability of the protein molecular structural parameters investigated.
Acknowledgements
This research has been supported by the grants from the Natural Sciences and Engineering Research Council of Canada (Individual Discovery Grant, Canadian federal government), the Ministry of Agriculture Strategic Feed Research Chair Programme, the Agricultural Bioproducts Innovation Programme of Agriculture and Agric-Food Canada, Beef Cattle Research Council and the Saskatchewan Agricultural Development Fund. W. G. N. O. provided assistance in conventional chemical and nutrient analyses. P. Y. was the leading investigator for the project. He designed the project, performed the studies, quantified amide I to amide II ratio and α-helix to β-sheet peak intensity ratio, performed statistical analyses, interpretated the data and wrote the manuscript. The authors thank D. Damiran and Z. Niu for providing assistance in sample collection, chemical analysis and spectral data collection. The authors state that there is no conflict of interest.






