Introduction
Cell-free DNA (cfDNA) in the human serum had been described before the structure of DNA was known (Ref. Reference Mandel and Metais1). It was thought that the cfDNA in serum is only the result of cell damage during coagulation of blood (Ref. Reference Steinman2). CfDNA was almost undetectable in plasma of healthy volunteers in the era before polymerase chain reaction (PCR). Presence of cfDNA was seen as a pathological sign. The immunoelectrophoresis assays used to detect DNA at that time had shown that cfDNA appeared in the circulation during haemodialysis, likely because of the damage of leucocytes (Refs Reference Steinman and Ackad3, Reference Fournie4). It took decades until scientists recognised that cfDNA might be of interest for diagnosis, monitoring, and even understanding the pathogenesis of various diseases. Later studies showed that cfDNA is also present in plasma under physiological conditions in healthy volunteers (Ref. Reference Zhong5). Because the clearance of cfDNA is very fast (between 4 and 30 min) (Refs Reference Rumore6, Reference Garcia Moreira7), it might become a useful biomarker for monitoring of rapid changes in a patient (Ref. Reference Avriel8). This is further supported by the fact that a simple fluorometric assay or real-time PCR is sufficient for its quantification. Moreover, rather than pure quantification, detection of different targets in DNA enables detection of cfDNA from different sources. For example, cfDNA containing regions specific for Y-chromosomes proved useful as a biomarker of graft rejection in female recipients of male donor organs (Ref. Reference Lui9). Other strategies included the detection of single-nucleotide polymorphism (SNP) genotyping of cfDNA from donor and recipient (Ref. Reference Snyder10), or detection of DNA mutations specific for tumours (Ref. Reference Fleischhacker and Schmidt11). Using DNA sequencing, especially massive parallel sequencing, the sensitivity and specificity have improved (Ref. Reference De Vlaminck12). Even though costs for such high-throughput analyses decrease, fast, simple and low-cost analyses are most promising strategies for clinical practice and include methods such as targeted end-point PCR combined with quantitative PCR or molecular bar-codes.
This review describes the basics of cfDNA biology, the use of cfDNA as a biomarker for renal tissue damage and summarises the data on the role of cfDNA in kidney disease pathogenesis.
Biological characteristics of cfDNA
The origin of cfDNA
CfDNA is released from cells by apoptosis, necrosis and netosis – production of extracellular traps by neutrophils, which actively release DNA-histones complexes to kill pathogens (Ref. Reference Allam13) (Fig. 1). These mechanisms are associated with various diseases, but occur also under physiological conditions (Ref. Reference Breitbach, Tug and Simon14). The principal cells that release cfDNA are blood cells. This has been demonstrated in patients after sex-mismatched bone marrow transplantation. In women, nearly 60% of plasma cfDNA originated from male donor-derived haematopoietic cells. On the other hand, a multivariate analysis in patients on haemodialysis showed that an important determinant of cfDNA is the blood pressure suggesting that vascular injury might contribute to the pool of plasma DNA in some patients (Ref. Reference Jeong da15).
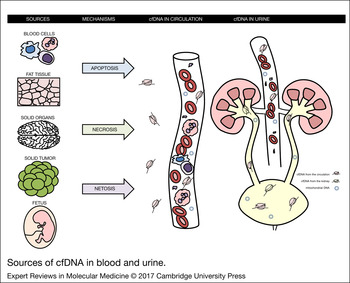
Figure 1. Sources of cfDNA in blood and urine. Plasma cfDNA is mostly derived from blood cells. An alternative source are tissue cells, fat cells, especially in obese patients, tumour cells in the patients with solid tumours and placental cells in pregnant women. The main mechanisms involved in the release of cfDNA include apoptosis, necrosis and netosis. Urine contains cfDNA derived from plasma.
After transplantation of solid organs such as liver, heart and kidney, the cfDNA from the donor accounts only for a very small (<1%) or an even undetectable portion of the whole plasma cfDNA, confirming its main origin from haematopoietic cells (Ref. Reference Lui16). Using specific extraction kits for isolation and detection of cfDNA and massive parallel sequencing, even such small amounts of cfDNA can be sufficient for the diagnostic purposes, for example, as shown for acute rejection after heart (Ref. Reference De Vlaminck12) and kidney transplantation (Ref. Reference Beck17). In pregnant women foetal cfDNA appears in maternal plasma (Ref. Reference Lo and Chiu18). Obesity is another factor contributing to cfDNA quantity. Continuous turnover of fat cells leads to release of cfDNA and a twofold higher concentration than in lean subjects (Ref. Reference Haghiac19).
The size of cfDNA
Fragments of cfDNA are 185–200 bases long in plasma of healthy volunteers, while the length of cfDNA fragments released from malignant cells shows higher variability (Refs Reference Chan20, Reference Schwarzenbach, Hoon and Pantel21). It is present in form of nucleoprotein complexes. The size of the fragments of cfDNA is important. When large amounts of small fragments (160–200 bases) of single-stranded DNA (ssDNA) were injected into mice, despite low blood levels, the DNA was found in the glomeruli even after 24 h (Ref. Reference Carlson22). The opposite results, that is, high blood levels but no presence in glomeruli, were observed for larger fragments of 2–6 kb (Ref. Reference Carlson22). These results indicate that the plasma cfDNA might be trapped in the glomeruli based on its size by a yet unknown mechanism. However, in the mentioned experiment ssDNA was used which is different from standard human double-stranded DNA. In addition, naked DNA was applied making it difficult to interpret the information for physiological and pathophysiological conditions where DNA is mostly bound to proteins. In pregnant women, maternal and fetal cfDNA are circulating in plasma. Both are fragmented, but fetal DNA is overrepresented in the slightly lower fragment sizes, which can be helpful for amplification of fetal DNA and enable further analyses (Ref. Reference Li23). Similarly, on lupus fragments of cfDNA in plasma are much shorter than in healthy probands (Ref. Reference Chan24).
Haemodialysis was proposed as a model to study the biology of cfDNA (Ref. Reference Rumore6). During haemodialysis, cfDNA is released into the circulation and resembles the cfDNA found in other diseases. Interestingly, after cessation of haemodialysis, the half-life of the produced cfDNA is only 4 min suggesting a high turnover rate (Ref. Reference Rumore6). Deoxyribonuclease activity is inversely proportional to the concentration of cfDNA. Low deoxyribonuclease activity in plasma could be the cause of increased concentration of cfDNA in some diseases. In cancer patients, low deoxyribonuclease activity might contribute to increase in cfDNA fragments size (Refs Reference Cherepanova25, Reference Stephan26). The highest deoxyribonuclease activity among all tested tissues and body fluids was found in urine. The enzyme activity in the kidney is higher than in most analysed organs (Ref. Reference Nadano, Yasuda and Kishi27). This could partially explain the very low concentration and high fragmentation of cfDNA in the urine. The physiological or pathological role of deoxyribonuclease activity in the kidney is, however, not clear.
Clearance of cfDNA
How cfDNA is cleared from plasma is unknown. Liver was shown to play an important role in the trapping and clearance of DNA, chromatin or mononucleosomes injected into mice (Refs Reference Gauthier, Tyler and Mannik28, Reference Emlen and Mannik29, Reference Du Clos30). Plasma nucleases were found to only partially contribute to the degradation of plasma cfDNA in humans (clearance of fetal cfDNA in maternal plasma) (Ref. Reference Lo31) or injected DNA in mice (Ref. Reference Chused, Steinberg and Talal32). This was further supported by a genome-wide association study, where the concentration of cfDNA was not associated with polymorphisms of the deoxyribonuclease gene, but with polymorphisms of genes in the UDP-glucuronosyltransferase 1 family, especially UGT1A1 (Ref. Reference Jylhava33). The enzyme encoded by the UGT1A1 locus is important for glucuronidation – a major pathway of detoxification of xenobiotics, but also a number of endogenous substances (Ref. Reference Williams34). The findings of this association study require confirmation and yet lack mechanistic explanation, but point to a likely involvement of the liver and the reticuloendothelial system in cfDNA clearance.
Patients with chronic renal failure do not have a higher cfDNA concentration in plasma despite the relatively low molecular weight of cfDNA (Ref. Reference Korabecna35). This indicates that kidneys might not be important for cfDNA clearance, potentially because of the negative charge of DNA. Also in animal experiments, the DNA uptake in kidneys was minimal (Ref. Reference Gauthier, Tyler and Mannik28). However, urine contains plasma-derived cfDNA. Kinetics of fetal cfDNA in the maternal circulation and in urine was assessed in women after delivery (Ref. Reference Yu36). While in the first hours after birth the clearance of fetal DNA was rapid, during the next hours it slowed down. After 2 days, fetal cfDNA was not detectable in maternal blood anymore. This study suggests that although kidney might be involved in clearance of cfDNA, this seems to be only a minor effect compared with yet unknown extrarenal clearance mechanisms. Whether this is also true for patients suffering from acute or chronic renal failure is unknown.
Sources of cfDNA and cfDNA as a biomarker
Tumours as a source of cfDNA
The concentration of cfDNA in plasma is increased in cancer patients and might serve as a potential tumour marker (Ref. Reference Anker37). One reason might be the reduced deoxyribonuclease activity in plasma of cancer patients (Ref. Reference Tamkovich38). Another, more likely reason, is that a part of the cfDNA in plasma is derived from tumour tissue cells undergoing apoptosis or necrosis (Ref. Reference Papadopoulou39) (Fig. 1). Cancer induces netosis and the produced extracellular traps lead to damage of the endothelium and the supplied organs (Ref. Reference Cedervall40). Other studies have brought results by showing that the interindividual variability in plasma cfDNA quantity and integrity is much higher than the difference between healthy controls and prostate cancer patients (Ref. Reference Boddy41). Higher cfDNA integrity in plasma was found in patients with hepatocellular carcinoma suggesting that the association might be tumour type-dependent (Ref. Reference Chen42). It might also be influenced by the analysis used. When ALU gene was used for the analysis of DNA integrity it was able to distinguish prostate cancer from benign prostate hyperplasia (Ref. Reference Feng43). Tumour cfDNA can be detected in the urine and it does not have to be derived from tumours of the urinary tract (as will be discussed later). Even in patients with colorectal carcinoma, the typical K-ras mutations can be detected in cfDNA isolated from urine (Ref. Reference Su44). At least in some study, the mutations in cfDNA were detected with much better sensitivity in urine compared with plasma or serum (95% versus 40% versus 35%). Sequence analysis and epigenetic analysis, but not pure quantification of cfDNA, will more likely be of use for cancer diagnosis and monitoring as shown in several important studies in the last years (Refs Reference Chan45, Reference Chan46, Reference Thierry47). CfDNA analysis from plasma or urine has proven to be a suitable alternative to tissue analysis for testing BRAF mutations prior to targeted therapy. The tested patient population was small, but the results were consistent with DNA analysis from the tumour tissue at 83% (Ref. Reference Janku48). In recent studies, similar results were found in a much larger set of gastric cancer patients where the utility of urine and plasma samples for screening and monitoring of gene alterations was tested. Epidermal growth factor receptor (EGFR) mutations were correctly determined by urinary and plasma cfDNA analysis in 92 and 99%, respectively (Ref. Reference Shi49). The ability to track EGFR mutations in the urine was also tested for lung cancer patients showing that urinary cfDNA is comparably informative to plasma cfDNA and tumour tissue analysis, while it is clear that urine is much easier to obtain and so, a very valuable source of tumour DNA applicable for clinical monitoring of the disease progress and treatment effects (Ref. Reference Chen50). CfDNA measured in plasma of patients with renal cell carcinoma was on average eightfold higher than in healthy controls. It decreased after nephrectomy and represented an independent prognostic factor, as it was associated with the risk of tumour reoccurrence (Refs Reference Perego51, Reference de Martino52, Reference Wan53). Analysis of microsatellite loss of heterozygosity detected in plasma cfDNA could predict the prognosis of the patients. CfDNA in patients with renal cell carcinoma had a much higher integrity of cfDNA compared with healthy controls. When longer target sequences (400 bp and more) were amplified by PCR, only cfDNA from patients was positive suggesting that the cfDNA derived from tumour cells is less fragmented (Ref. Reference Gang54). The reason might be that in contrast to other cfDNA, tumour-derived cfDNA likely originates from necrotic and not apoptotic cells (Refs Reference Hauser55, Reference Shaked56). However, a recent study focusing on the abundance of short and long fragments in patients with renal cell carcinoma found the complete opposite result – longer fragments were less abundant in patients in comparison to controls (Ref. Reference Lu57). These inconsistencies clearly require more research.
CfDNA can be used to study the methylation status of various tumour-related genes. In most of the patients with renal cell carcinoma, hypermethylated genes were found in the cfDNA (Refs Reference de Martino52, Reference Hauser58). The response of renal cell carcinoma to therapy can also be monitored by analysis of cfDNA, with high concentrations being associated with worse outcome (Ref. Reference Feng59). Circulating mitochondrial cfDNA was found to be elevated in patients with renal cell carcinoma, prostate and bladder cancer. In the latter two also the integrity of mitochondrial DNA was higher indicating that mitochondrial cfDNA might be a marker of tumours in the urinary tract (Ref. Reference Ellinger60) (Fig. 2).

Figure 2. Kidney injury and cfDNA. CfDNA from tumours can be detected in the plasma and urine of patients with renal cell carcinoma. During the kidney injury, a larger amount of mitochondrial cfDNA and fragmented cfDNA is present in the urine. The plasma cfDNA and transrenal urinary cfDNA (i.e. plasma-derived), can be analysed using real-time PCR and sequencing for screening, diagnosis and monitoring of disease progression.
Analysis of the urinary DNA and its methylation status can reveal information about the malignant processes in the kidney. Promoter hypermethylation of several tumour suppressor genes was found to be similar in serum, urine and the tumour in patients with renal cancer (Refs Reference Hoque61, Reference Urakami62, Reference Hoque63). In patients with bladder cancer, microsatellite analysis (serum and urinary cfDNA) and PCR detecting of specific mutations (urinary cfDNA) revealed associations of DNA alternations with the disease (Refs Reference von Knobloch64, Reference Szarvas65, Reference Little66). Even a simple quantification of urinary cfDNA could help to screen patients with bladder cancer. In patients with bladder cancer urinary cfDNA is higher than in healthy controls (on average nearly threefold using Picogreen detection, and 14-fold higher using real-time PCR targeting a larger DNA fragment) (Ref. Reference Chang67). A follow-up study using other methods of urinary cfDNA quantification (spectrophotometry, fluorometry and real-time PCR) failed to confirm the results in another cohort of bladder cancer patients (Ref. Reference Zancan68). Similarly to circulating cfDNA, longer fragments and, thus, the higher integrity of urinary cfDNA was associated with bladder and prostate cancer (Refs Reference Casadio69, Reference Casadio70). In contrast, lower integrity index of urinary cell-free RNA was found in patients with renal cell carcinoma (Ref. Reference Zhao71). It was thought that changes in urinary DNA integrity could also be used as an early noninvasive diagnostic marker for prostate cancer. However, both sensitivity and specificity of integrity analysis have been shown to be lower than for prostate-specific antigen levels measurement (Ref. Reference Salvi72).
Transplants as a source of cfDNA
Donor DNA in plasma of transplant recipients was found by detecting the Y chromosome in plasma of women receiving the kidney or liver from male donor (Ref. Reference Lo73) and the presence of donor DNA in body fluids of the recipient was confirmed in a later study (Ref. Reference Zhong74). The detection of donor DNA can be based on Y chromosome DNA quantification, HLA differences, but also on insertion-deletion polymorphisms that are easy to detect using real-time PCR (Ref. Reference Adamek75). Elevation of transplant-derived DNA sequences in urine was associated with graft rejection in rats (Ref. Reference Martins76). Donor DNA was found in the plasma of the recipient after kidney transplantation and was higher in HLA mismatched donor–recipient pairs without immunosuppressive treatment. In a clinical study monitoring 100 patients during three months after transplantation, total cfDNA and not donor DNA or urinary DNA could be used for the detection of acute graft rejection episodes (Refs Reference Garcia Moreira77, Reference Sigdel and Sarwal78), which might have been because of the low number of patients with sex-mismatched transplants included in the study. CfDNA was analysed in plasma and urine samples using the digital PCR method to monitor organ rejection after kidney transplantation. Donor-derived cfDNA could only be detectable in urine samples, but unlike serum creatinine analysis, cfDNA analysis could not monitor or predict clinical development after kidney transplantation (Ref. Reference Lee79). Missing correlation between the donor-kidney contribution to the cfDNA in urine and serum creatinine values in stable patients was found also using SNP polymorphisms analysis (Ref. Reference Cheng80). Recently, the diagnostic usability of circulating cfDNA analysis was shown for the monitoring of the transplanted kidney. The status of the donor kidney is usually assessed using histological analysis of biopsy specimens. Renal biopsy cannot be done as often as blood sampling. In this study, the donor-derived cfDNA fraction of 1% from total cfDNA was determined as a threshold of organ damage meaning that donor-derived cfDNA above 1% shows very likely a rejection of the kidney (Ref. Reference Bloom81). As late detection of graft rejection is a major clinical problem, the analysis of cfDNA could help to solve this issue, thus warranting further research in this area. Especially, as a dynamic analysis of donor DNA found that it gets higher in plasma of the recipient sooner than other biochemical parameters (Ref. Reference Beck17) and much sooner than any histological markers can be found (Ref. Reference Gielis82). The early diagnosis of graft rejection achievable with cfDNA analysis enables early and, thus, more effective interventions that improve the prognosis of patients (Ref. Reference Oellerich83).
Tissue damage as a source of cfDNA
CfDNA in plasma is elevated in a number of pathologies associated with tissue damage, necrosis or apoptosis. Mere quantification of plasma cfDNA is a very nonspecific marker, yet in some pathologies the concentrations of cfDNA are extremely high. Concentrations of cfDNA in patients with myocardial infarction increase by more than 50% and correlate with prognosis (Refs Reference Antonatos84, Reference Destouni85). This has been confirmed later in a similar cohort using a quick fluorometric assay to quantify cfDNA, in which the cfDNA correlated with other known myocardial damage markers (Ref. Reference Shimony86) and was associated with patient outcome (Refs Reference Huang87, Reference Gornik88, Reference Zaravinos89). Similarly, in an animal model of stroke, cfDNA was higher after 24 h and the concentration was associated with the infarct volume (Ref. Reference Boyko90). In both rats and patients, cfDNA increases after traumatic brain injury and the subsequent rapid decrease is associated with a better survival (Refs Reference Macher91, Reference Ohayon92). Similar results were seen in patients with other types of trauma (Ref. Reference Ren93). CfDNA is also higher in patients with burns and correlates with the severity of the tissue damage and prognosis (Refs Reference Chiu94, Reference Shoham, Krieger and Perry95). In general, no matter which organ is affected, the amount of cfDNA in plasma seems to correlate with the volume of damaged tissue and in some cases with the prognosis.
CfDNA is associated with inflammatory markers in nonagenarians (Ref. Reference Jylhava96). The increase in the quantity of cfDNA in plasma is not restricted to local injuries, but has been studied also in systemic diseases. Plasma cfDNA is elevated fourfold in rheumatoid arthritis (Ref. Reference Zhong97) and correlates with the apnoea–hypopnoea index in patients suffering from sleep apnoea syndrome (Ref. Reference Shin98). Even more, it decreases when sleep apnoea is treated using continuous positive airway pressure along with markers of inflammation and oxidative stress (Ref. Reference Ye99). CfDNA is higher in diabetic patients with and without microvascular complications in comparison to controls (Ref. Reference El Tarhouny100) and in various acute disorders such as sepsis and respiratory failure (Refs Reference Arnalich, Lopez-Collazo and Montiel101, Reference Okkonen102). Unsurprisingly, a recent study showed that cfDNA and nucleosomes are higher also in patients with haemolytic uremic syndrome very likely because of the activation of neutrophils (Ref. Reference Ramos103). In ANCA-associated vasculitis the situation is not that clear. While some studies have shown higher cfDNA (Refs Reference Yoshida104, Reference Nakazawa105), in other the disease was not associated with higher cfDNA, nucleosomes or neutrophil extracellular traps (Ref. Reference Wang106). In summary, cfDNA is elevated in various diseases and its mere quantification is a very unspecific, but in some instances very sensitive biomarker. Owing to its high turnover and rapid clearance (Ref. Reference Lo31), it is suitable for monitoring dynamics of tissue damage and/or recovery. Before monitoring of cfDNA will be applicable for clinical practice, much more has to be learned about the metabolism of cfDNA.
Inflammation and cfDNA
PCR analysis using primers designed for the human genome does not allow detecting sequences from other organisms. In 21% of patients on haemodialysis, bacterial cfDNA was found in the blood despite negative cultures (Ref. Reference Bossola107). The presence of bacterial cfDNA in plasma is associated with slightly higher concentrations of interleukin 6 and C-reactive protein, similarly to patients with peritoneal dialysis (Ref. Reference Kwan108). The low-grade inflammation is very likely mediated by Toll-like receptor 9 that recognises CpG islands in the bacterial DNA (Ref. Reference Hemmi109). Other DNA sensors such as the stimulator of interferon genes (STING) might also be involved (Ref. Reference Lood110). Mitochondrial DNA resembles bacterial DNA and in some pathologies it was found to be elevated in plasma (Ref. Reference Zachariah111). This is likely to be because of tissue damage that might lead to activation of the immune system and potentially to sepsis-like syndrome (Ref. Reference Zhang112). Elevated mitochondrial cfDNA is associated with high mortality of patients in the intensive care unit (Ref. Reference Nakahira113). Whether these associations are causal remains to be elucidated. At least in lupus it was recently shown that mitochondrial DNA induced immune system activation, especially, when it was oxidised (Ref. Reference Lood110). It can only be speculated whether this is of importance for other cfDNA-associated disorders such as sepsis, obesity or liver failure.
Analysis of plasma cfDNA derived from mitochondria and bacteria has been suggested as a potential tool to distinguish septic from sterile causes of systemic inflammatory response syndrome (Ref. Reference Sursal114). This is important because also in sepsis with bacteraemia, the total human cfDNA is elevated, especially in patients with a worse prognosis (Refs Reference Huttunen115, Reference Forsblom116). At least in one study in sepsis patients, concentrations of cfDNA gave a better prognostic information than the standard clinical scoring systems or established coagulation and inflammatory markers (Ref. Reference Dwivedi117). In another study, higher cfDNA was a prognostic factor for patients with sepsis-induced acute kidney injury (Ref. Reference Clementi118). Analysis of cfDNA, especially when done quickly and easily, could improve the decision making in critical care patients (Ref. Reference Avriel8). This shows that the rapid turnover of cfDNA is advantageous for monitoring acute pathologic processes and could also be used for other fields, not only emergency medicine.
Urinary cfDNA as a biomarker
The cfDNA present in the urine might originate from plasma. Since it passes through the glomerular filter, it was termed transrenal DNA (Ref. Reference Botezatu119). When the PCR is designed to target very short amplicons and contamination is prevented, the transrenal urinary cfDNA can be used for the detection of fetal gender in pregnant women. However, the specificity of the assay is much lower than for plasma cfDNA (Ref. Reference Shekhtman120). Using bisulphite sequencing it was found that cfDNA in the urine originates from blood cells, kidney and urinary tract cells and the proportion of cfDNA from these cell types is very variable (Ref. Reference Cheng80). It has been shown that after haematopoietic stem cell transplantation, donor-derived DNA can be found in the urine of the recipient. In contrast to the urinary recipient cfDNA, the donor-derived urinary cfDNA was less fragmented than in plasma suggesting another source of this urinary cfDNA than plasma, potentially the donor-derived cells of the renal tubular system (Ref. Reference Hung121). Urinary tract infections lead to an increase in urinary cfDNA confirming that the urinary tract might be an important source of urinary cfDNA (Ref. Reference Garcia Moreira122). The currently available data suggest that the contribution to the pool of urinary cfDNA from plasma and from the urinary tract vary depending on the analysed disease and yet unknown factors.
Injection of lipopolysaccharide or other toxins in experimental animals leads to an increase in cfDNA concentrations in both, plasma and urine (Ref. Reference Bret123). The urinary cfDNA was proposed as a marker of kidney damage, as it increases in an animal model studying the nephrotoxicity of mercury (Ref. Reference Bret124). Heat-shock proteins and cfDNA in the urine have been suggested to be a possible indicator of acute renal injury in sepsis, but clinical studies brought rather sobering results (Ref. Reference Vaara125). Another urinary marker derived from DNA is 8-oxo-7,8-dihydro-2′-deoxyguanosine. This is often used for the assessment of oxidative DNA damage or subsequent DNA repair, but its validity has been questioned in the past (Ref. Reference Cooke, Henderson and Evans126) and might not be related to urinary cfDNA. In patients with hantavirus-induced nephropathy, plasma cfDNA, but not the urinary cfDNA, was associated with clinical symptoms (Ref. Reference Outinen127). In a recently published case report about a patient with otitis media, the cfDNA in the urine was used for the detection of Mycobacterium tuberculosis causing the infection (Ref. Reference Petrucci128). The determination of cfDNA amount in urine, similarly to plasma cfDNA, is unspecific and increases in a number of different diseases, but combining quantitation and targeted analyses of the sequences can increase its diagnostic value.
Recent studies pointed to the possibility that urinary cfDNA could be a valuable biomarker if the analysis targets specifically mitochondrial DNA in the urine. The concentration of urinary mitochondrial DNA was higher in patients with acute kidney injury. Even more, it was related to the progression of their disease. This finding was confirmed in an animal experiment where the ischaemia-induced renal injury and the urinary mitochondrial DNA correlated positively (Ref. Reference Whitaker129). Mitochondrial DNA in the urine is higher in patients with chronic renal injury as well. When patients with essential and renovascular hypertension were compared with healthy volunteers, urinary mitochondrial DNA was higher in two independent cohorts. It was even associated with standard markers of renal injury such as kidney injury molecule-1 and neutrophil gelatinase-associated lipocalin (Ref. Reference Eirin130). Further results focus not only on the quantity, but also on the quality and fragmentation status of urinary mitochondrial DNA can be expected.
The role of cfDNA in disease pathology
Systemic lupus
Systemic lupus erythematosus was the first disease that was thought to be caused by cfDNA. First, it was hypothesised that the immunogenic stimulus that induces the production of auto-antibodies is the ssDNA (Ref. Reference Koffler131). Later, it has been shown that an injection of lipopolysaccharide into mice leads to release of DNA that causes the immune reaction with auto-antibody production. The complexes of DNA with the particular antibodies were found in the glomerular basement membrane where they induced sterile inflammation (Ref. Reference Izui132). The DNA bound to the antibodies had a low molecular weight suggesting a high fragmentation (Ref. Reference Bruneau and Benveniste133). The presence of DNA in the immune complexes seems to be specific for lupus and not for other types of nephritis (Refs Reference Williams and Adu134, Reference Adu, Dobson and Williams135). However, when plasma cfDNA was measured using more modern techniques, the concentrations were only slightly higher in lupus patients and showed a considerable overlap with control subjects (Ref. Reference McCoubrey-Hoyer, Okarma and Holman136). It becomes clear that cfDNA is not a single factor involved in the pathogenesis of lupus. By analysing DNA from necrotic and apoptotic cells administered to mice it was shown that the resulting DNA fragments were short and very likely protected from further degradation by histones in form of nucleosomes (Ref. Reference Pisetsky and Jiang137). It could be that cfDNA has to be bound to histones or other molecules or be released in a specific context to induce auto-inflammation. This context could be represented by cell death that is accompanied by the release of damage-associated molecular patterns that drive inflammation. This can lead locally to the death of further cells. This vicious cycle might be an important pathomechanism in several autoimmune diseases including lupus, graft rejection and cancer (Ref. Reference Linkermann138).
The most important work describing cfDNA in lupus patients uses massively parallel sequencing of plasma DNA (Ref. Reference Chan24). The results clearly showed that the cfDNA in lupus patients has a skewed size distribution of cfDNA fragments with a higher proportion of shorter fragments. The shortening even correlated with disease activity. It seems that the short fragments are protected by binding to immunoglobulins. In addition, the methylation analysis revealed that these short fragments are hypomethylated in comparison to cfDNA that is not protected by immunoglubulins. The cfDNA in lupus patients is not only a biomarker. Current therapeutic efforts in lupus are focusing on the clearance of neutrophil extracellular traps and, thus, on decreasing of circulating cfDNA, using deoxyribonuclease (Refs Reference Barnado, Crofford and Oates139, Reference Gupta and Kaplan140).
Lupus patients cannot efficiently degrade neutrophil extracellular traps because of lower deoxyribonuclease activity (Ref. Reference Hakkim141). This might be because of genetic variability as a polymorphism in the deoxyribonuclease gene was found to be related to the risk for lupus nephritis (Ref. Reference Camicia142). Low deoxyribonuclease activity seems to be a predisposing factor for lupus (Ref. Reference Gajic-Veljic143). Besides genetic factors, the activity of deoxyribonuclease in plasma can be inhibited by the presence of the complement protein C1q (Ref. Reference Leffler144). Conversely, the neutrophil extracellular traps activate complement. The starting mechanism of this circulus vitiosus is unknown, but the resulting higher amounts of extracellular traps lead to endothelial dysfunction and potentially also to renal injury (Refs Reference Allam13, Reference Carmona-Rivera145). Interestingly, indices for a similar mechanism have been found in ANCA-associated microscopic polyangiitis (Refs Reference Yoshida104, Reference Nakazawa105), but this is not generally accepted and some studies have brought opposite results (Ref. Reference Wang106). It is proposed that cfDNA is released via netosis and that the subsequent induction auto-antibody production, immune complexes formation and inflammation associated cell death including netosis further increase cfDNA (Ref. Reference Fenton146). This cfDNA includes mitochondrial DNA that seems to be immunogenic and leads to the production of antibodies (Ref. Reference Wang147). Interestingly, administration of recombinant deoxyribonuclease did not affect the disease activity – neither in mice (Ref. Reference Verthelyi148) nor in patients with lupus (Ref. Reference Davis149). However, the enzymatic activity might have been inhibited by circulating factors in plasma. Whether targeting pathways recognising cfDNA, that is, Toll-like receptors, or use of deoxyribonucleases that are not inhibited by factors present in the plasma of lupus patients, is a relevant clinical option that remains to be tested (Ref. Reference Ahn, Ruiz and Barber150). The only clinical possibility to inhibit netosis is to apply anti-inflammatory drugs, which already are a mainstay therapy in lupus patients (Ref. Reference Lapponi151).
Kidney injury
A recently published experiment analysed the role of cfDNA in acute kidney injury induced by ischaemia. The renal injury was associated with histological damage, negative functional consequences and higher intra-renal DNA debris. More importantly, administration of exogenous deoxyribonuclease ameliorated the induced damage and resulted in improvement of renal perfusion and functions (Ref. Reference Peer152). This indicates that cfDNA is not only a consequence of renal tissue damage, but its release can lead to an amplification of the damaging processes. It is not clear whether the positive effect of deoxyribonuclease is restricted to this specific model or it could be observed in other models of kidney failure. When mitochondrial DNA was analysed, its concentrations in the urine were found to be a good biomarker of acute as well as chronic renal injury (Refs Reference Whitaker129, Reference Eirin130). But beyond that, animal experiments proved its pathogenic role in the development of hepatic injury as a consequence of ischaemia-reperfusion injury of the kidney (Ref. Reference Bakker153). The released cfDNA, especially from mitochondria is recognised by the Toll-like receptor 9 and induces further damage to distant organs. This was proved by the analysis of the consequences of ischaemia-reperfusion injury of the kidney in Toll-like receptor 9 knock-out mice. While the renal injury was similar to the wild-type mice, the induced secondary hepatic injury was reduced by the genetic defect (Ref. Reference Bakker153). As mitochondrial DNA is not protected by histones, this is another potential therapeutic niche for deoxyribonuclease administration.
Circulating mitochondrial DNA was analysed in patients with systemic inflammatory response syndrome and subsequent acute kidney injury. Circulating mitochondrial DNA increases in this inflammatory disease, but does not correlate with renal damage and does not increase in acute kidney injury, but the amount of mitochondrial DNA in urine correlates with urinary markers of inflammation and coagulation and probably has a role in the pathophysiology of renal failure during systemic inflammatory response syndrome (Ref. Reference Jansen154). The pathophysiological effect of mitochondrial DNA in the development of acute renal failure during sepsis was described in the animal model. During sepsis the increased circulating mitochondrial DNA activates Toll-like receptor 9 and the signalling pathway leading to cytokine production, inflammation, oxidative stress and renal failure (Ref. Reference Tsuji155).
Haemodialysis
Haemodialysis increases the concentration of plasma DNA because of apoptosis of leukocytes as shown by the appearance of typical ladders after electrophoresis (Ref. Reference Atamaniuk156). Although the increase is observed in all patients, it varies quantitatively. Compared with concentrations before haemodialysis session, initially up to fourfold higher cfDNA concentrations are found during the procedure but decrease to the pre-dialyses levels within 30 min after haemodialysis (Ref. Reference Garcia Moreira7). The timing of sampling and cfDNA monitoring is important. One hour after haemodialysis no major difference to the basal values before haemodialysis was observed (Ref. Reference Cichota157). The temporary increase of cfDNA during haemodialysis did not correlate with either dialysis duration or its efficiency (Ref. Reference Opatrna158). Children on peritoneal dialysis have a twofold higher cfDNA when compared with healthy controls (Ref. Reference Ozkaya159) and the concentrations of cfDNA in the effluent are related to peritoneal membrane degeneration (Ref. Reference Pajek160) and peritonitis (Ref. Reference Virzi161). Haemodialysis induces inflammation and this can be monitored using cfDNA-based analyses of differences in methylation of promoters of pro-inflammatory genes (Ref. Reference Korabecna162). The released cfDNA itself could be the cause of the inflammation, but this requires further studies. One small study showed that the cfDNA concentration after haemodialysis was a predictor of all-cause mortality in these patients (Ref. Reference Tovbin163). A recent larger study focusing on mitochondrial DNA confirmed this finding. Patients in the highest quartile of plasma mitochondrial DNA had a much worse event-free survival than patients with lower mitochondrial DNA. In addition, the concentrations of mitochondrial DNA correlated positively with inflammatory markers (Ref. Reference Szeto164). Whether this represents a causal relationship is not clear.
Other diseases
Apart from its role in lupus, it is currently not clear whether cfDNA might also play a functional role in disease development or progression, in particularly those associated with very high cfDNA concentrations. In cancer, the release of tumour DNA into blood might possibly lead to the transformation of distant cells with the tumour-specific mutations via horizontal-transfer. After homologous recombination, these mutations could be functionally relevant and induce metastasis or secondary tumour formation at a different site (Ref. Reference Garcia-Olmo and Garcia-Olmo165). This hypothesis is called genometastasis and tries to explain how the transfer of tumour genes instead tumour cells could aid the spreading of solid tumours. The authors have shown that plasma of tumour-bearing rats contains tumour DNA (Ref. Reference Garcia-Olmo166), that is related to size of the tumour (Ref. Reference Garcia-Olmo167) and that this DNA can be internalised by cells in vitro (Ref. Reference Garcia-Olmo168). These experiments, however, await confirmation by other laboratories. Although the idea is simple and exciting, especially with regards to the potential new therapeutic modalities, further studies are needed to confirm the results. Whether cfDNA has any role in physiology and pathology of diseases, and whether administration of deoxyribonucleases has any therapeutic potential similar to ribonucleases that prevent atherosclerosis induced by extracellular RNA remains unclear (Refs Reference Simsekyilmaz169, Reference Fischer170).
In healthy elderly people cfDNA is associated with inflammation (Ref. Reference Jylhava171). In cell culture, cfDNA, but also plasma from patients after haemodialysis induces interleukin 6 production in human monocytes, suggesting a pro-inflammatory effect (Ref. Reference Atamaniuk172). On the other hand, a recently published experiment in mice suggests that injection of cfDNA isolated from mice with colitis into healthy mice prevents the subsequently induced colon inflammation (Ref. Reference Muzes173). In a murine lupus model, it was shown that the recognition of cfDNA by macrophages could be facilitated by the chromatin protein and inflammation marker high mobility group box 1 (Ref. Reference Li174). On the other hand, elevated deoxyribonuclease was found to be associated with kidney injury in urolithiasis (Ref. Reference Yusof175). These contradictory findings clearly show that more research on the effects of cfDNA is needed.
Technical issues
The concentration of cfDNA is very low and it is present only in very small fragments. This means that the isolation of cfDNA is not trivial and various isolation procedures have been tested with variable outcomes (Refs Reference Jorgez176, Reference Kirsch177, Reference Xue178, Reference Sharma, Vouros and Glick179). As a result of the physicochemical properties of cfDNA, either specific DNA isolation kits or a large volume of starting plasma or urine are required. For simple cfDNA measurement, venous plasma can be substituted by finger capillary plasma as shown in an experiment on exercising athletes (Ref. Reference Breitbach180). Serum has a 14-fold higher cfDNA concentration than plasma. Higher amounts are mainly caused by lysis of the donor cells during coagulation (Ref. Reference Lui9). Both serum and plasma are routinely used and suitable for the analyses (Ref. Reference Fleischhacker and Schmidt11), however, some cancer-related mutations might be undetectable in serum because of possible contamination by DNA released during coagulation (Ref. Reference Hibi181).
Analysis of cfDNA in body fluids, such as plasma or urine, as the so-called liquid biopsy has one major advantage over standard biopsy – noninvasiveness. The true clinical usefulness has yet to be confirmed in direct head to head comparisons with currently used standard tests. Beyond the quantity and integrity of cfDNA it will likely be of importance to describe the origin of cfDNA by methylation-specific markers. Protocols for the bisulphite conversion and subsequent PCR or sequencing analysis have been recently modified by using targeted amplification to allow analysis of samples with very low amount of cfDNA, including plasma and urine (Ref. Reference Jung182).
Automated cfDNA isolation protocols can help to increase the throughput of the measurement and also lower the risk of contamination (Ref. Reference Huang183). The total amount of cfDNA might be influenced by haemolysis during blood collection, storage or centrifugation. It has been shown that an additional centrifugation step after plasma separation is needed to exclude contamination with large cellular DNA (Ref. Reference Page184). Specific blood collection tubes that prevent cell lysis and, thus, contamination of cfDNA with genomic DNA as well as extracellular RNA from cytoplasmic RNA from lysed leucocytes are available, but increase the cost of sampling (Refs Reference Fernando185, Reference Hidestrand186, Reference Norton187, Reference Qin, Williams and Fernando188). The important factors of the pre-analytical phase that could potentially bias cfDNA analysis in plasma have been reviewed recently (Ref. Reference El Messaoudi189). If haemolysis and contamination are prevented, then plasma can be stored for several years at −20°C without major effects on cfDNA (Ref. Reference Koide190). Efforts are made to prepare a simple detection and quantification method of cfDNA without the need of DNA isolation or PCR. A fluorometric assay that quantifies cfDNA in serum is available, but its use for plasma samples was limited because of insufficient sensitivity (Ref. Reference Goldshtein, Hausmann and Douvdevani191). Currently available rapid assays based on a similar principle can be helpful for critically ill patients despite the lack of specificity (Ref. Reference Shoham, Krieger and Perry95). For other purposes, the gold standard is the real-time PCR targeting various genes. The choice of the target can, however, bias the outcome (Ref. Reference Devonshire192). The use of several target genes for the quantification of cfDNA improves the reliability of the analysis. Specific methods can be applied to quantify the cfDNA released in the form of neutrophil extracellular traps (Ref. Reference Kraaij193).
Apart from technical issues, before sampling, biological features should be taken into account. CfDNA seems not to be influenced by age (except of nonagenarians where cfDNA was higher), gender, menstrual cycle or frequency of blood donations (Refs Reference Zhong194, Reference Polcher195, Reference Jylhava196). This might be an advantage for a biomarker as it can be used for monitoring of rapid changes in a patient. Smoking increases cfDNA nearly threefold (Ref. Reference Urato197) and physical activity can temporarily increase cfDNA concentrations up to tenfold, at least for a few hours (Refs Reference Atamaniuk198, Reference Beiter199). One study reported relatively high intraindividual fluctuation of cfDNA concentrations in healthy individuals (13.5-fold) (Ref. Reference Zhong5).
Conclusion
CfDNA circulating in plasma or present in urine can be a useful source of information for prenatal diagnosis of genetic diseases, haemodialysis induced cell damage, renal or bladder tumour screening and monitoring, and the detection of kidney transplant rejection. With the use of modern sequencing techniques, the genome of the foetus or the tumour can be reconstructed and used for further genetic analyses. Several questions remain to be answered, for example, what is the role of kidney in the clearance of cfDNA from plasma? Or whether cfDNA might be also implicated in other renal diseases beyond lupus nephritis? What is the potential of cfDNA in nontumourous kidney diseases beyond transplant rejection? Finally, it should be mentioned that as other types of DNA, also cfDNA encodes genetic information, could be involved in rapid communication between cells in the form of horizontal gene transfer and might be a potential mechanism of action making it an even more exciting area of research (Refs Reference Peters and Pretorius200, Reference Vlkova201).
Acknowledgements
The authors are supported by grants from the German Research Foundation (DFG: SFB/TRR57 and SFB/TRR219; BO3755/3-1 and BO 3755/6-1 to P.B.), German Ministry of Education and Research (BMBF: STOP-FSGS-01GM1518A to P.B.), Slovak Research & Development Agency (APVV-16-0273 to P.C.) and Ministry of Education of the Slovak Republic (VEGA 1/0156/17 to B.V. and VEGA 1/0092/17 to P.C.).
Conflict of interest
The authors confirm that there are no conflicts of interest.




