Bacterial isolates from a clearcut outbreak of disease are frequently designated as a clone. The term clone is used in a wider context as proposed by Orskov & Orskov [Reference Orskov and Orskov1] ‘to denote bacterial cultures isolated independently from different sources, in different locations, and perhaps at different times but showing so many identical phenotypic and genetic traits that the most likely explanation for this identity is a common origin’. Apparently, such clones have features that make them successful and relatively stable over prolonged periods. Strains of these clones are notorious for their involvement in colonization and infection, epidemic spread and accumulation of antibiotic resistance genes.
Within Acinetobacter baumannii, three major epidemic multidrug-resistant (MDR) clones have been identified in European isolates and, hence, were named European (EU) clones I–III [Reference Dijkshoorn2, Reference van Dessel3]. Since their initial description, it has been well documented that these clones occur worldwide and consensus has been to rename them International clones (IC) I–III [Reference Diancourt4, Reference Zarrilli5].
Various methods have been used to identify isolates to these clones, including amplified fragment length polymorphism (AFLP) analysis [Reference Dijkshoorn2, Reference van Dessel3, Reference van den Broek6], multiplex PCR and multilocus sequence typing (MLST) of housekeeping genes. Results of the latter method can be compared between laboratories to provide insight into the global epidemiology of A. baumannii. A complete agreement of clone identification by AFLP analysis and MLST of seven housekeeping genes was observed [Reference Diancourt4]. Isolates identified by AFLP to IC clones I–III correspond to clonal complexes, named CC1–3, respectively. Furthermore, a few tentative novel clonal complexes were distinguished in this study, i.e. CC10, CC15 and CC32.
Recently, epidemic isolates from wounded US military service members were described as belonging to a novel clone which had previously been identified in isolates of the Leiden AFLP database [Reference Petersen7]. Two isolates of this clone were identified as CC32 by MLST [Reference Diancourt4]. The objective of the present study was to characterize this clone in more detail.
Twelve non-replicate clinical isolates, collected between 1980 and 2005 from five hospitals in five European countries, one hospital in Kuwait and by the US military healthcare system, were studied (Table 1). They were selected to represent a cluster of 31 clinical isolates in the Leiden AFLP database, which comprises fingerprints from >1500 Acinetobacter spp. isolates obtained from 1984 to 2011 [Reference van den Broek6]. Sixteen isolates of this cluster were unrelated in time and space, while a set of 15 isolates (different patients) was from the outbreak among the US soldiers [Reference Petersen7]. The 12 isolates of study clustered together at 81·2%, indicative for clonal relatedness. Definitive clone allocation was done by MLST [Reference Diancourt4].
Table 1. Characteristics of the Acinetobacter baumannii strains studied
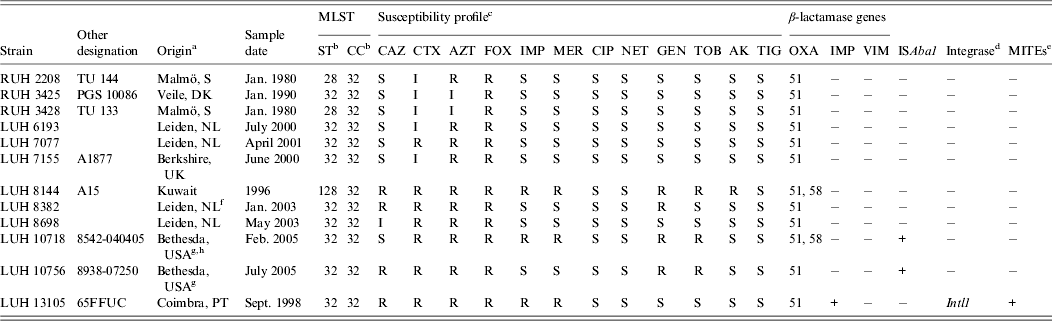
a PT, Portugal; S, Sweden; DK, Denmark; NL; The Netherlands.
b ST, Sequence type; CC clonal complex (according to Institute Pasteur database, Diancourt et al. [Reference Diancourt4]).
c CAZ, ceftazidime; CTX, cefotaxime; AZT, aztreonam; FOX, cefoxitin; IMP, impenem; MER, meropenem; CIP, ciprofloxacin; NET, netilmicin; GEN, gentamicin; TOB, tobramycin; AK, amikacin; TIG, tigecycline.
d PCR screening of Intl1, Intl2 and Intl3.
e MITEs, Miniature inverted transposable elements.
f Patient transferred from Egypt.
g Patient transferred from Iraq (12).
h Outbreak related (12).
Antimicrobial susceptibility was tested by disc diffusion for ceftazidime, cefotaxime, aztreonam, cefoxitin, imipenem, meropenem, ciprofloxacin, netilmicin, gentamicin, tobramycin, amikacin and tigecycline (Oxoid, UK). Results were interpreted according to CLSI (2010) and EUCAST (2013) guidelines. Multiplex polymerase chain reaction (PCR) was used for screening OXA-type β-lactamases. PCR was used to screen for IMP and VIM carbapenemases, integrases Intl1, Intl2, Intl3 [Reference Domingues, Nielsen and Da8], ISAba1 and miniature inverted transposable elements (MITEs) [Reference Domingues9].
The grouping of the 12 strains of the novel clone on the basis of AFLP analysis and their comparison with reference strains of IC I–III, all identified by MLST, is shown in Figure 1. The clones are well-separated with a linkage level each of ⩾80%, except for IC II which is more diverse (clustering at 72·3%). AFLP analysis can also be used for strain identification at a cut-off level of 90% [Reference Dijkshoorn2, Reference van den Broek6]. At this level, nine different types could be distinguished, against only three sequence types by MLST. By MLST, isolates were identified to clonal complex (CC32) with sequence types ST32 (n = 9), ST28 (n = 2) and ST128 (n = 1) (Table 1).

Fig. 1. Amplified fragment length polymorphism (AFLP) tree novel EU clone IV and condensed branches of clones I–III with isolates from the Leiden AFLP database. Strains of each cluster have been characterized by multi-locus sequence typing and were unrelated in time and/or space of origin [Reference Metan11]. Values at nodes represent distance of strains within each cluster. ST, Sequence type, CC, clonal complex. Numbers of strains are in parentheses.
Antibiotic susceptibility was variable in the isolates. Three isolates were carbapenem resistant. One included the 65FFC isolate with a MITE-borne integron carrying the IMP-5 determinant [Reference Domingues, Nielsen and Da8]. The same 439-bp MITE-like structure was found to be associated with class 1 integrons carrying metallo-β-lactamases in diverse clinical Acinetobacter spp. strains [Reference Domingues9]. All others were negative for IMP, VIM, integrases and MITEs. The bla OXA-58-type carbapenemase was identified in the other two carbapenem-resistant isolates, one from US soldiers and one from Kuwait. The gene bla OXA-51-type which is usually present in A. baumannii was detected in all isolates. Carbapenem resistance of these two isolates could be associated with the OXA-58 gene. Only two isolates, LUH10718 and LUH10756, carried ISAba1, the latter not resistant to carbapenems (Table 1). ISAba1 was not found in the carbapenem-resistant LUH8144. Nevertheless, the presence of other insertion sequences, not screened in this study, may explain the resistance. Markedly, this strain was susceptible to ceftazidime. A similar phenotype (imipenem non-susceptible and susceptible to ceftazidime) was recently reported in an OXA-58 producer ST23 A. baumannii from Mozambique [Reference Bonnin10].
Clonal lineages have long been associated with multiple drug resistance, and this feature might confer a selective advantage for their expansion. IC II is the most marked example of success of worldwide dissemination, which has been associated with the acquisition of carbapenem resistance by diverse sublineages of this clone. Although this clone appears to be prevalent, MDR strains belonging to IC I continue to be reported in some regions. In a recent study, blood isolates from a Turkish hospital were found to belong to another novel international clone, CC15 with most organisms being positive to OXA-58 genes and showing carbapenem resistance [Reference Metan11]. Moreover, eight clonal lineages of carbapenem-non-susceptible A. baumannii were delineated, mostly based in rep-PCR typing method, originated from multiple geographical locations, and thus, referred as worldwide clonal lineages (WW1–8) [Reference Higgins12]. According to recent recommendations, some of the CC32 isolates of the current study can be classified as MDR since they show resistance to antibiotics in more than three antimicrobial categories (Table 1).
The total number of the representatives of the described novel clone (CC32) is relatively limited. However, the occurrence of epidemic-related isolates, some from Leiden, The Netherlands, and others from the US military healthcare system, underscore their association with outbreaks/cross-infections. These criteria, the finding of carbapenem resistance and the collection at different times and locations indicates the epidemic potential of this clone.
Since the discovery of the first isolates of CC32 was in Europe, it might be plausible to designate this new clone as European clone IV. However, due to the increasing international consensus to abandon the use of the term EU clones I, II and III in favour of International clones I, II and III due to their worldwide dissemination [Reference Diancourt4, Reference Zarrilli5], CC32 could be considered to represent an emerging International clone IV. Future studies are needed to clarify the extension of CC32 spread and to determine if it warrants classification as IC IV or some other designation. In the meantime, A. baumannii clone terminology remains confusing, since it is based on diverse typing methods and/or origin/spread of isolates, a process of international harmonization is desperately needed and should be a priority of experts in the field.
ACKNOWLEDGEMENTS
This work received partial financial support from the Center of Pharmaceutical Studies, University of Coimbra, Portugal. N. Mendonça and S. Domingues are supported by grants SFRH/BPD/45815/2008 and SFRH/BD/49061/2008, respectively, from the Fundação para a Ciência e a Tecnologia, Lisbon, Portugal.
Dr Petersen is an employee of the US government. The opinions and assertions contained herein are of the authors and are not to be construed as official or reflecting the views of the Department of the Navy, or the U.S. Government.
DECLARATION OF INTEREST
None.




