Introduction
Market research shows consumers believe organic foods are safer and healthier than conventional foods (Sofos, Reference Sofos2008; Umberger et al., Reference Umberger, Boxall and Lacy2009). Comparisons of organic and conventional foods have shown greater concentrations of beneficial polyphenolic cancer-fighting compounds and a lower incidence of pesticide residues in organic vs conventional foods (Barański et al., Reference Barański, Średnicka-Tober, Volakakis, Seal, Sanderson, Stewart, Benbrook, Biavati, Markellou, Giotis, Gromadzka-Ostrowska, Rembiałkowska, Skwarło-Sońta, Tahvonen, Janovská, Niggli, Nicot and Leifert2014). However, some organic foods have been linked to food-borne illness outbreaks and microbial safety protocols for organic foods may need further evaluation to minimize the risk of food-borne outbreaks (Harvey et al., Reference Harvey, Zakhour and Gould2016). Escherichia coli O157:H7 and Salmonella spp. are the major pathogens associated with organic foods and responsible for 33 and 44% of all outbreaks, respectively (Harvey et al., Reference Harvey, Zakhour and Gould2016). These two pathogens are particularly economically damaging and were estimated to cost the industry over $12 billion in 2010 in the United States alone (Scharff, Reference Scharff2012). From 1998 to 2008, there were a total of 7246 viral- or bacterial-vectored food-borne outbreaks (Crowe et al., Reference Crowe, Mahon, Vieira and Gould2015), with 16 of the last 18 total bacterial and viral outbreaks associated with organic foods occurring between 2005 and 2015 (Harvey et al., Reference Harvey, Zakhour and Gould2016). More recently, studies have shown equal or lower contamination in organic foods compared with conventional counterparts (Gad et al., Reference Gad, Abo-Shama, Harclerode and Fakhr2018; Merlini et al., Reference Merlini, Pena, da Cunha, de Oliveira, Rostagno and Antunes2018; Becker et al., Reference Becker, Stoll, Schlz, Kulling and Huch2019).
Since 2005, the overall rate of organic sales has increased, with a 29.3% increase in meat and poultry sales (Sustainable Food News, 2016). This trend is expected to continue to into the future (Scaglia, Reference Scaglia2017), and studies showing greater nutritional quality in organic vs conventional beef (Średnicka-Tober et al., Reference Średnicka-Tober, Barański, Seal, Sanderson, Benbrook, Steinshamn, Gromadzka-Ostrowska, Rembiałkowska, Skwarło-Sońta, Eyre, Cozzi, Krogh-Larsen, Jordon, Niggli, Sakowski, Calder, Burdge, Sotiraki, Stefanakis, Yolcu, Stergiadis, Chatzidimitriou, Butler, Stewart and Leifert2016; Ribas-Agusti et al., Reference Ribas-Agusti, Diaz, Sarraga, Garcia-Regueiro and Castellari2019) may bolster sales. However, cattle are known reservoirs for E. coli and Salmonella spp. and often carry them asymptomatically, thus making beef an excellent entry point for pathogens into the food supply (Hsi et al., Reference Hsi, Ebel, Williams, Golden and Schlosser2015). It is therefore imperative to determine the prevalence of these pathogens in cattle-based systems and develop effective intervention protocols to ensure food safety compliance.
Since synthetic fertilizers are prohibited in organic systems (USDA-AMS-NOP, 2018), raw livestock manure is often applied to enhance soil fertility. However, manure can harbor E. coli O157:H7 and Salmonella spp. as a vehicle for transmission of enteric pathogens (Kudva et al., Reference Kudva, Blanch and Hovde1998; Islam et al., Reference Islam, Morgan, Doyle, Phatak, Millner and Jiang2004; Franz et al., Reference Franz, Van Diepeningen, De Vos and Van Bruggen2005; Holley et al., Reference Holley, Walkty, Blank, Tenuta, Ominski, Krause and Ng2007). The survival rate of these pathogens is highly variable and depends on climatic factors (Muirhead, Reference Muirhead2009). Contaminated manure has been shown to effectively transfer pathogens to vegetation (Holley et al., Reference Holley, Walkty, Blank, Tenuta, Ominski, Krause and Ng2007; Jacobsen and Bech, Reference Jacobsen and Bech2012). Thus, the application of manure can increase the risk of contamination events, particularly as vegetation may provide an oral pathogen inoculation for grazing cattle.
Livestock feed ingredients such as dried forage, grains, hay and silage have been shown to be a source of both E. coli O157:H7 and Salmonella spp. (Davies and Wales, Reference Davies and Wales2013). Under supportive environmental conditions these organisms can undergo replication, and, thus, should be monitored in the farm environment (Krytenburg et al., Reference Krytenburg, Hancock, Rice, Besser, Gay and Gay1998; Lynn et al., Reference Lynn, Hancock, Besser, Harrison, Rice, Stewart and Rowan1998; Dargatz et al., Reference Dargatz, Strohmeyer, Morley, Hyatt and Salman2005; Wales et al., Reference Wales, Allen and Davies2010). Examples of food-borne outbreaks associated with animal feed include Salmonella enterica subtype Agona that was traced to contaminate fish meal that was fed to cattle (Clark et al., Reference Clark, Kaufman, Gangarosa and Thompson1973). Davis et al. (Reference Davis, Hancock, Rice, Call, Digiacomo, Samadpour and Besser2003) found that strains of Salmonella spp. and E. coli O157:H7, isolated from feed, could be found in environmental and fecal samples as well.
Recently, interest in integrating crops and livestock on organic farms has generated concern for adherence to food safety rules related to manure application and contamination in the United States (USDA-FDA, 2018). In one study evaluating the effectiveness of the USDA-NOP 120-day raw manure set-back rule, Patterson et al. (Reference Patterson, Navarro-Gonzalez, Jay-Russell, Aminabadi, Antaki-Zukoski and Pires2018) found low concentrations of E. coli in soil 84–111 days after sheep grazing in an integrated system. In our study, a model integrated crop–livestock system, consisting of annual rotations between crops and forage/pasture systems, was established in three states to measure crop and animal performance, in addition to food safety aspects. The objectives of the study described here were to determine the prevalence of E. coli O157:H7 and Salmonella spp. in feed, fecal, hide and meat samples from cattle raised in these integrated systems in Iowa, Minnesota and Pennsylvania from August 2015 to March 2017. While the original hypothesis was that there would not be any contamination issues in the integrated system, the alternative hypothesis of differences in E. coli and Salmonella spp. prevalence rates between months of the study and sample types within these systems would be useful in developing site-specific on-farm and processing facilities' food safety protocols.
Materials and methods
Experimental design and sampling overview
Model integrated systems were established as observational study sites at the University of Minnesota West Central Research and Outreach Center (Morris, MN), the Rodale Institute (Kutztown, PA) and an on-farm site in Iowa (Erlandson Farm, Greenfield, IA). Specific details of the model system design and grazing regime are described in Phillips et al. (Reference Phillips, Heins, Delate and Turnbull2017). Briefly, 3-year integrated system rotations were established within 10- to 50-ha sites in Fall 2015–Spring 2016 that included small grain forages of rye (Secale cereale L.) and wheat (Triticum aestivum L.) for grazing the first year, followed by row crops of soybeans (Glycine max L.) and corn (Zea mays L.), and then a mixed-species pasture of fescue (Festuca arundinacea L.), orchardgrass (Dactylis glomerata L.), red clover (Trifolium pratense L.) and white clover (Trifolium repens L.) (Fig. 1). The Iowa site began with the pasture year followed by a second year of small grain forage, due to land constraints. The study began in August 2015 with 6 to 7-month-old cattle (organic, mixed dairy breed herd consisting of Holstein, Montbeliarde, Viking Red, Normande and Jersey genetics). Each site selected local, organic herds to enhance the site specificity of the project. Sampled herd sizes were 4, 12 and 11 head for Iowa, Pennsylvania and Minnesota, respectively. Cattle grazed on forages/pasture beginning in April when forages were at least 15 cm tall until slaughter in October 2016 in Minnesota and Iowa, and from April to October 2016 in Pennsylvania (cattle not harvested until 2017). Crops and cattle were raised according to USDA-AMS organic regulations as set forth in the National Organic Program (NOP) rules (USDA-AMS-NOP, 2018). Pastures (IA site) and small grain forages (MN and PA sites) received no fertilization, outside of manure deposits, during the study. If pasture/forage was insufficient for cattle nutritional needs, certified organic commercial feed was provided to the cattle based on recommendations from an organic livestock specialist at the University of Minnesota. Cattle had access to minerals and water ad libitum throughout the study.

Fig. 1. Experimental design showing the two crop sequence (CS) treatments (rotations) replicated three times as 3-acre plots. The rotations consisted of wheat or rye for grazing in Yr 1, followed by corn or soybean in Yr 2 and ending with a return to pasture in Yr 3–4. Each 3-acre plot was sub-divided into four 0.75-acre paddocks during the grazing phases of the study.
Feed and forage sampling
Commercial bagged feed and live forage/pasture samples were obtained from each site and tested for presence of E. coli O157:H7 and Salmonella spp. For feed samples, 100 g of feed were randomly sampled with sterile equipment from feed bags. Forage/pasture matter was clipped at the base of the plant, within a square-foot quadrat, in five random locations of the forage/pasture. Forage samples included plant foliage, along with incidental roots and soil matter. Feed consisted of organic grains, or an organic mixed ration with mineral supplements. Feed and forage/pasture were sampled throughout the season and when steers were moved onto new pastures. Samples were placed in sterile collection bags with cold packs that did not come into direct contact with the samples to avoid influencing microbial analysis. Samples were shipped overnight to the Department of Food Science and Human Nutrition, Iowa State University (Ames, IA) for analysis. Samples were stored in a refrigerator (4°C) and processed within 12 h of arrival. The number of samples taken was based on the number of different feed types provided to the cattle in the study.
Collection of fecal samples
Fecal samples were collected from freshly deposited cow pats at the start of the study at each site, before harvest, and, whenever there was a change in feed. Sterile sample collection cups were utilized to store and transport fecal samples. Fecal sample cups were shipped with cold packs as described above to the Department of Food Science and Human Nutrition at Iowa State University and stored and processed as described above. The number of samples taken was based on the number of cattle present at that specific location.
Hide and meat sampling
Hide surface samples were collected prior to harvest and meat steaks were collected after harvest, using a 10 × 10-cm2 template with an EZ Reach™ sponge sampler (World Bioproducts, IL, USA). The rump area was sampled for hide and a loin section was swabbed on both sides for the meat samples. All samples were shipped with cold packs as described above to the Department of Food Science and Human Nutrition at Iowa State University and stored and processed as described above. The number of samples taken was based on the number of cattle present at that specific location.
Microbial analysis
Sub-samples (25 g) of feed/forage samples from each site were analyzed for microbial content. Fecal samples were analyzed as sub-samples of 10 g each. For meat swabs, 1 ml of sampling buffer was extruded and sampled. Samples were then selectively enriched in parallel for E. coli O157:H7 or Salmonella spp. and analyzed using miniVidas® kits (bioMerieux® SA, Marcy-l'Etoile, France) and processed as per manufacturer's instructions. Samples that tested positive were then subjected to confirmatory methods as prescribed by USDA and FDA BAM standards (USDA-FDA, 2017).
E. coli O157:H7 colonies were identified by plating onto Sorbitol MacConkey® agar (Difco®, BD, Franklin Lakes, NJ, USA) with Cefixime® and Tellurite® (CT-SMAC) (Oxiod Microbiological Products, Hampshire, UK) as colorless colonies. Colorless colonies were considered typical and 3–4 colonies per plate, if present, were selected at random to be analyzed using a Dry Spot® latex kit for E. coli O157 (Oxiod Microbiological Products, Hampshire, UK). Agglutination with the latex reagents was considered as positive for E. coli O157:H7.
Samples that were presumptively positive for Salmonella spp. were streaked in duplicate onto Brilliant green sulfa agar (BGS) (Difco®, BD, Franklin Lakes, NJ, USA) and xylose lysine tergitol™ 4 agar (XLT-4) (Difco®, BD, Franklin Lakes, NJ, USA). Typical colonies were selected as per manufacturer's instructions. Colonies were then used to inoculate butts of triple sugar iron and lysine-iron agar which were then incubated with loosened caps at 35 ± 2°C for 24 h, and examined as a set, for positive responses in accordance with the manufacturer's guidelines.
Statistical analysis
Data were analyzed by the type of sample (feed/forage, fecal, hide and meat), month and location using PROC FREQ and ANOVA in SAS/STAT® 9.3. Percentages displayed in Results are the number of positive or negative samples over the total number of samples obtained from that subset of data.
Results
Prevalence of pathogens in feed/forage samples
The weather during the study period across the three locations was typical of each state's climate, which is categorized as a humid continental climate. Warm to hot and humid weather occurred during the main grazing period of May to September, with the warmest months from June to August (Table 1).
Table 1. Weather conditions in 2016 at the three sites of the ‘Integrated Organic Crops and Livestock’ project

A total of 53 feed/forage samples across all locations was tested for the presence of E. coli O157:H7 and Salmonella spp. over the course of the study. Overall, five samples tested positive for E. coli O157:H7 and one sample was positive for Salmonella spp. (Tables 2 and 3). No E. coli O157:H7 or Salmonella spp. were detected in feed samples from Iowa. Three of sixteen forage samples from Minnesota tested positive for E. coli O157:H7, which were obtained from pastures in June and July 2016, 2 months prior to cattle harvest. Two of the twenty feed samples from Pennsylvania tested positive for E. coli O157:H7 from the mixed ration feed in August. The sole positive Salmonella spp. sample in feed was from Pennsylvania (Table 2) and also was obtained in August from the mixed ration feed. All positive Salmonella spp. samples were isolated in the warmer months of June, July and August.
Table 2. Prevalence of Salmonella spp. in feed, fecal and hide samples of cattle in the ‘Integrated Organic Crops and Livestock’ project

a Sample not taken in this month.
Table 3. Prevalence of E. coli O157:H7 in feed, fecal and hide samples of cattle in the ‘Integrated Organic Crops and Livestock’ project

a Sample not taken in this month.
Prevalence of pathogens in fecal samples
A total of 124 fecal samples across all sites was analyzed throughout the study. Nine samples tested positive for E. coli O157:H7 and four tested positive for Salmonella spp. (Tables 2 and 3). All positive samples for Salmonella spp. were from Iowa in August, prior to cattle harvest. E. coli O157:H7 was isolated from one sample from Minnesota, obtained during the first sampling period in August at the post-weaning stage. Eight fecal samples from Pennsylvania tested positive for E. coli O157:H7 (Table 3). While all Iowa fecal samples tested negative for E. coli O157:H7 (Table 3), the Pennsylvania samples that tested positive, collected prior to cattle harvest, caused a significant difference between months sampled, due to low numbers from the other sites (Table 5).
Prevalence of pathogens in hide and meat samples
Sampling of hides occurred prior to cattle harvest. Forty-three hide samples were obtained in July and August 2016 from Minnesota and in August 2016 from the Iowa herd. There were no hide samples from Pennsylvania because harvesting did not occur until the next year when cattle market prices were higher. E. coli O157:H7 was not detected on any hide samples, while Salmonella spp. was detected on eight samples obtained in August (Tables 2 and 3). Minnesota and Iowa cattle each had four positive samples for Salmonella spp. The prevalence rate for Salmonella spp. was significantly greater on hides compared with other sample types (Table 4) and greater prevalence occurred in August compared with the other months sampled (Table 5).
Table 4. Prevalence of E. coli O157:H7 and Salmonella spp. by sample type across the three study sites (MN, IA and PA) in the ‘Integrated Organic Crops and Livestock’ project

1Means followed by the same letter down the column are not significantly different at P ≤ 0.05.
Table 5. Prevalence of E. coli O157:H7 and Salmonella spp. by month across all sample types (feed, fecal and hide) from the three study sites (MN, IA, PA) in the ‘Integrated Organic Crops and Livestock’ project
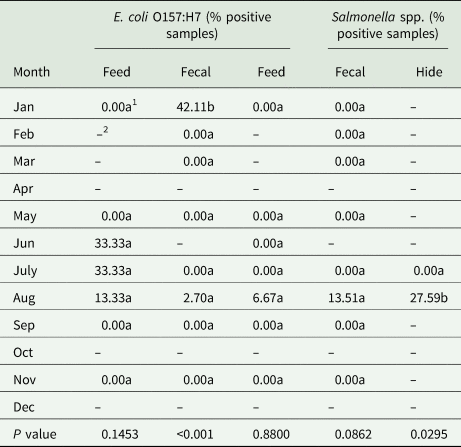
1Means followed by the same letter down the column are not significantly different at P ≤ 0.05.
2Sample not taken in this month.
Neither E. coli O157:H7 nor Salmonella spp. was detected from any meat sample obtained from study animals (Tables 2 and 3). Again, no meat analysis occurred in Pennsylvania due to the postponed harvesting. Due to the greater number of positive fecal samples in Pennsylvania over the course of the study, that site expressed a greater rate than the other two sites (Table 6), but the E. coli prevalence rate was within the range reported by Reinstein et al. (Reference Reinstein, Fox, Shi, Alam, Renter and Nagaraja2009).
Table 6. Prevalence of E. coli O157:H7 and Salmonella spp. in fecal, feed, hide and meat samples by study sites (MN, IA and PA) in the ‘Integrated Organic Crops and Livestock’ project

1Means followed by the same letter down the column are not significantly different at P ≤ 0.05.
2Sample not taken at this location.
Discussion
Feed/forage pathogen prevalence
The greatest number of positive E. coli and Salmonella spp. feed/forage samples was obtained in the warmer months of June, July and August in Pennsylvania and Minnesota, which were the months frequently associated with higher pathogen prevalence rates in the United States, Finland and Australia (Barkocy-Gallagher et al., Reference Barkocy-Gallagher, Arthur, Rivera-Betancourt, Nou, Shackelford, Wheeler, Koohmaraie and Hruska2003; Lahti et al., Reference Lahti, Ruoho, Rantala, Hänninen and Honkanen-Buzalski2003; Hussein, Reference Hussein2007; Williams et al., Reference Williams, Ward, Dhungyel and Hall2015). Salmonella spp. was detected in 1.89% of the 53 samples in this study, which was lower than that determined by Krytenburg et al. (Reference Krytenburg, Hancock, Rice, Besser, Gay and Gay1998), who detected Salmonella spp. in 9.8% of feed samples consisting of several feed components, obtained between July and November, from dairy farms and feedlots. Davis et al. (Reference Davis, Hancock, Rice, Call, Digiacomo, Samadpour and Besser2003) isolated S. enterica in 0.8% of feed samples, obtained from conventional dairy farms in Washington and northwest Idaho, consisting of apple waste, fish meal, hay, grains, cotton seed, rye pellets, beet pulp and pea flour. Over an 8-yr period (2002–2009), Li et al. (Reference Li, Bethune, Jia, Lovell, Proescholdt, Benz, Schell, Kaplan and McChesney2012) reported a 12.5% Salmonella contamination rate in feeds. E. coli O157:H7 was found in three pasture samples from Minnesota and two feed samples consisting of a total mixed ration in Pennsylvania. Overall, the 9.43% E. coli O157:H7 prevalence rate was greater than that found in the previous study by Davis et al. (Reference Davis, Hancock, Rice, Call, Digiacomo, Samadpour and Besser2003), but less than the 30.1% rate reported by Lynn et al. (Reference Lynn, Hancock, Besser, Harrison, Rice, Stewart and Rowan1998) in dairy cattle feed samples. The E. coli prevalence rate observed in this study, however, was similar to the 14.9% prevalence rate observed by Sargeant et al. (Reference Sargeant, Sanderson, Smith and Griffin2004) in feed bins from feedlots.
Fecal pathogen prevalence
Of the 124 fecal samples over the course of the study, E. coli O157:H7 was isolated from eight samples from Pennsylvania in January and one from Minnesota in August. This prevalence rate of 7.26% was similar to the average rate of 9.3%, and within the range of 0–24.4%, found by Reinstein et al. (Reference Reinstein, Fox, Shi, Alam, Renter and Nagaraja2009) from fecal samples of organically raised cattle. Jacob et al. (Reference Jacob, Fox, Drouillard, Renter and Nagaraja2009) found a lower prevalence rate of E. coli O157:H7 (5.1%) in an experimental setting where cows were fed dried distillers grains or dry rolled corn. Cho et al. (Reference Cho, Bender, Diez-Gonzalez, Fossler, Hedberg, Kaneene, Ruegg, Warnick and Wells2006) found an equivalent E. coli O157:H7 rate of 7.4% at an organic dairy farm in Minnesota. A study of grass-fed cattle in Australia demonstrated a prevalence rate of 10% (Fegan et al., Reference Fegan, Vanderlinde, Higgs and Desmarchelier2004a). These studies, however, used molecular techniques for enrichment which may have provided a more sensitive evaluation than typically evaluated (Eriksson and Aspan Reference Eriksson and Aspan2007).
Salmonella spp. prevalence rates were similar to previous studies of organic and conventional production systems. The overall Salmonella spp. prevalence rate of 3.26% was similar to reports from Minnesota and Wisconsin where Salmonella spp. was detected in 5.2% of fecal samples collected from organic dairy farms (Fossler et al., Reference Fossler, Wells, Kaneene, Ruegg, Warnick, Eberly, Godden, Halbert, Campbell, Bolin and Zwald2005). Similarly, in a study conducted with feedlot cattle in Nebraska, Salmonella spp. prevalence was determined to be 5.4% (Schmidt et al., Reference Schmidt, Agga, Bosilevac, Brichta-Harhay, Shackelford, Wang, Wheeler and Arthur2015). Feces of grass-fed beef cattle in Australia were found to have a Salmonella spp. prevalence rate of 4.5% (Fegan et al., Reference Fegan, Vanderlinde, Higgs and Desmarchelier2004b). Herd size has been found to be a significant factor on farms with higher prevalence rates (Fossler et al., Reference Fossler, Wells, Kaneene, Ruegg, Warnick, Eberly, Godden, Halbert, Campbell, Bolin and Zwald2005; Habing et al., Reference Habing, Lombard, Kopral, Dargatz and Kaneene2012). Thus, smaller herds dispersed across a larger grazing area may lead to lower Salmonella spp. prevalence rates compared with feedlot conditions.
Hide pathogen prevalence
Of the 43 hide samples from cattle in Iowa and Minnesota collected before harvest, no samples tested positive for E. coli O157:H7. However, four hide samples from Iowa and four from Minnesota tested positive for the presence of Salmonella spp. The overall prevalence rate for Salmonella spp. on hides was 18.6%. Most studies isolated higher rates of E. coli O157:H7 (30–60%) from hides during harvest (Barkocy-Gallagher et al., Reference Barkocy-Gallagher, Arthur, Rivera-Betancourt, Nou, Shackelford, Wheeler, Koohmaraie and Hruska2003; Arthur et al., Reference Arthur, Bosilevac, Brichta-Harhay, Kalchayanand, Shackelford, Wheeler and Koohmaraie2007; Brichta-Harhay et al., Reference Brichta-Harhay, Guerini, Arthur, Bosilevac, Kalchayanand, Shackelford, Wheeler and Koohmaraie2008). Barkocy-Gallagher et al. (Reference Barkocy-Gallagher, Arthur, Rivera-Betancourt, Nou, Shackelford, Wheeler, Koohmaraie and Hruska2003) determined positive E. coli O157:H7 samples on 60.6% of hides in a commercial beef processing plant that sourced cattle from the Midwest, while Brichta-Harhay et al. (Reference Brichta-Harhay, Guerini, Arthur, Bosilevac, Kalchayanand, Shackelford, Wheeler and Koohmaraie2008) examined hides of cattle from processing facilities across the United States and obtained 33.3% positive hide samples. The current results are in agreement with a study conducted on feedlot cattle in Texas, where a decrease in E. coli O157:H7 was observed on hides after transport and before harvest (Barham et al., Reference Barham, Barham, Johnson, Allen, Blanton and Miller2002).
Salmonella spp. prevalence was higher in hide samples (18.6%) taken before harvest than in fecal samples (3.33%), a pattern observed by both Barham et al. (Reference Barham, Barham, Johnson, Allen, Blanton and Miller2002) and Barkocy-Gallagher et al. (Reference Barkocy-Gallagher, Arthur, Rivera-Betancourt, Nou, Shackelford, Wheeler, Koohmaraie and Hruska2003), who reported an 89% rate (hide) vs 46% (fecal) (Barham et al., Reference Barham, Barham, Johnson, Allen, Blanton and Miller2002) and 71% (hide) vs 4.4% (fecal) (Barkocy-Gallagher et al., Reference Barkocy-Gallagher, Arthur, Rivera-Betancourt, Nou, Shackelford, Wheeler, Koohmaraie and Hruska2003). Greater variation in Salmonella spp. prevalence has been observed within seasons (Brichta-Harhay et al., Reference Brichta-Harhay, Guerini, Arthur, Bosilevac, Kalchayanand, Shackelford, Wheeler and Koohmaraie2008) and before/after transport (Barham et al., Reference Barham, Barham, Johnson, Allen, Blanton and Miller2002). The Salmonella spp. prevalence rate on hides observed in our study was much lower than other studies, which reported rates from 6 to 93.8% (Barham et al., Reference Barham, Barham, Johnson, Allen, Blanton and Miller2002; Barkocy-Gallagher et al., Reference Barkocy-Gallagher, Arthur, Rivera-Betancourt, Nou, Shackelford, Wheeler, Koohmaraie and Hruska2003; Arthur et al., Reference Arthur, Bosilevac, Brichta-Harhay, Kalchayanand, Shackelford, Wheeler and Koohmaraie2007; Brichta-Harhay et al., Reference Brichta-Harhay, Guerini, Arthur, Bosilevac, Kalchayanand, Shackelford, Wheeler and Koohmaraie2008) and were highly variable. An Australian study on Salmonella spp. prevalence found similar results, reporting that 2% of hides and 7% of fecal samples from organically raised cows were positive for Salmonella spp. (Fegan et al., Reference Fegan, Vanderlinde, Higgs and Desmarchelier2005). The Salmonella spp. prevalence rate, although low in comparison with most studies in organic and conventionally raised cattle, followed the reported trend of higher rates on hides than in fecal samples. It should be noted that a herbal fly repellent containing plant essential oils was used on cattle at all locations. Plant essential oils have been show to kill both E. coli O157:H7 and Salmonella species (Burt, Reference Burt2004; Turgis et al., Reference Turgis, Borsa, Millette, Salmieri and Lacroix2008; Bajpai et al., Reference Bajpai, Baek and Kang2012; Boskovic et al., Reference Boskovic, Zdravkovic, Ivanovic, Janjic, Djordjevic, Starcevic and Baltic2015; Khaleque et al., Reference Khaleque, Keya, Hasan, Hoque, Inatsu and Bari2016) and may have reduced pathogen load, but this result would need to be substantiated in future studies.
Meat pathogen prevalence
All meat swabs were negative for E. coli O157:H7 and Salmonella spp. However, all samples were taken post-chilling, which could considerably reduce the prevalence of both pathogens. Hauge et al. (Reference Hauge, Nesbakken, Moen, Røtterud, Dommersnes, Nesteng, Østensvik and Alvseike2015) saw a significant reduction (1–11%) post-chilling in total E. coli populations in their study of two Norwegian abattoirs, although there was no indication of whether the cattle were organically or conventionally raised. Similar to the 0% prevalence in this study, however, Fegan et al. (Reference Fegan, Vanderlinde, Higgs and Desmarchelier2005) also did not isolate any Salmonella spp. post-chilling in organically raised Australian cattle. The low pathogen prevalence rate from earlier sampling points in this study, and the interventions applied, could have led to a further decrease in the number of pathogens by the time of slaughter.
Conclusions
Of critical concern to consumers is the avoidance of human diseases caused by pathogens, such as E. coli O157:H7 and Salmonella spp., that could be vectored by livestock in integrated systems. The integrated crop–livestock system in this study demonstrated a high probability of meeting food safety goals for limiting E. coli O157:H7 and Salmonella spp. contamination in the forage, feed, feces, hide and meat of grass-based organic cattle. Overall, the study found a low prevalence rate of E. coli O157:H7 in feed and fecal samples that was similar to previous studies conducted with organic and conventional cattle. The E. coli O157:H7 rate of 9.43% in feed in this study was comparable with previous studies of feed bins in feedlots, but higher than previously reported for feed sampled directly from feed mills, suggesting that on-farm management strategies, such as ensuring feed bag and bin closing/coverage after each use, could reduce pathogen load. Salmonella spp. prevalence, however, at 1.89% of samples, was similar to feed samples from dairy farms in Washington and northwest Idaho, and lower than other studies. Feed from the months of June, July and August had a higher number of positive E. coli O157:H7 samples than other months, as warmer periods may be particularly vulnerable entry points for these pathogens into the food chain, thereby warranting strict food safety protocols.
Isolation of E. coli O157:H7 from 7.26% of fecal samples from study cattle was similar to that previously reported for organic dairy cows in Minnesota. The fecal Salmonella spp. prevalence rate of 3.26% was much lower than previous studies conducted in different production systems and may be associated with the smaller sized, more dispersed and grass-based herd compared with other studies. No E. coli O157:H7 was detected in hide samples taken prior to harvest, where previous studies indicated a higher prevalence rate on hides. The use of an essential-oil-based organic fly repellent may have served a dual purpose of insect and bacterial reduction, but further testing should occur before recommending for this use. Salmonella spp. prevalence on hides, with 18.6% testing positive, matched previous studies conducted at abattoirs. E. coli O157:H7 and Salmonella spp. were not isolated from any meat samples, which was also observed in a similar sized herd of Australian organic cattle. The previously documented pathogen reduction from chilling operations is likely associated with the lack of pathogens on any meat sample in this study. Further studies to identify exact sources of potential bacterial contamination along the production chain, from field to slaughter, will help increase the implementation of safe, integrated crop–livestock systems.










