Introduction
In the field of human-assisted reproductive technology, normal fertilization is confirmed by the presence of a two-pronuclear (2PN) zygote with two polar bodies (PBs) 16–20 h after insemination. Mono-pronuclear (1PN) zygote is defined as abnormal fertilization.
The utility of 1PN zygotes in in-vitro fertilization (IVF) has been a controversial topic for the past 20 years. It is generally advised against transferring 1PN embryos due to the possibility of chromosomal anomalies, even in cases where there are no viable 2PN embryos (De los et al., Reference De los, Apter and Coticchio2016; Azevedo et al., Reference Azevedo, Pinho and Silva2014; Yan et al., Reference Yan, Li and Shi2010; Reichman et al., Reference Reichman, Jackson and Racowsky2010). Nevertheless, recent studies indicate that predicting embryonic ploidy based solely on the number of pronuclei is not adequate (Lim et al., Reference Lim, Goh and Su2000; Bradley et al., Reference Bradley, Traversa and Hobson2017; Capalbo et al., Reference Capalbo, Treff and Cimadomo2017). Cytogenetic assessments have shown that the diploid rate of 1PN embryos can reach as high as 80.5% (Capalbo et al., Reference Capalbo, Treff and Cimadomo2017; Rosenbusch, Reference Rosenbusch2014). Numerous studies have demonstrated that the transfer of 1PN embryos can lead to the birth of healthy offspring (Si et al., Reference Si, Zhu and Lyu2019; Li et al., Reference Li, Huang and Zhuang2021).
Transfer of 1PN embryos can potentially benefit the patients when no other viable embryos are available. However, to date, there are no established protocols for the safe and efficacious transfer of 1PN embryos.
Pre-implantation genetic testing for aneuploidy (PGT-A) is an alternative for selecting diploid bi-parental 1PN embryos for transfer. Embryo biopsy, however, necessitates specialized equipment and extensive expertise. Moreover, the invasive procedure of this method poses a potential threat to subsequent embryonic development. These factors limit the universal application of PGT-A in 1PN embryo selection.
The time-lapse system facilitates the evaluation of embryonic development without altering the culture environment. It has been reported that some morphokinetic might be useful to predict ploidy in 2PN embryos (Bamford et al., Reference Bamford, Barrie and Montgomery2022). The pronuclear area and the diameter of 1PN zygotes are correlated with the subsequent developmental potential (Araki et al., Reference Araki, Itoi and Honnma2018). The 1PN embryos that reached the blastocyst stage exhibit similar morphokinetic features compared to 2PN zygotes (Mateo et al., Reference Mateo, Vidal and Carrasco2020). A recent study demonstrated that 1PN zygotes were formed when the fusion of the sperm took place within 18 μm from the point of the second polar body extrusion (Wei et al., Reference Wei, Enatsu and Furuhashi2022). However, the above studies did not explore the association between pronuclear characteristics, developmental potential, and the genetic constitution of 1PN embryos.
Therefore, we aim to investigate whether the pronuclear characteristics are related to the developmental potential and chromosomal constitution in 1PN embryos.
Methods
Design and subjects
Patients who underwent IVF or intracytoplasmic sperm injection (ICSI) in the Women and Children’s Hospital of Chongqing Medical University between December 2021 and September 2022 were included in this retrospective cohort study.
Only 1PN zygotes which were transferred to the time-lapse incubators on the oocyte retrieval day were included in this study because these zygotes could have intact embryo images. Those 1PN zygotes which were transferred to the time-lapse incubator the next day after insemination were excluded. In addition, zygotes temporarily displayed 1PN as follows were also excluded, including the asynchronous formation of 2PN, asynchronous disappearance of 2PN, a fusion of 2PN, and zygotes with one pronucleus and a micronucleus. Zygotes unsuitable for time-lapse assessment were not used to investigate the pronuclear characteristics due to excessive residual corona cells or inadequate orientation. Furthermore, fifteen 1PN zygotes were also excluded because they were confirmed as 2PN when the time-lapse images were retrospectively analyzed.
The primary outcomes were the blastocyst formation and good-quality blastocyst rates. The secondary outcomes were the pronuclear characteristics and genetic constitution of 1PN embryos.
A total of 388 1PN zygotes were used to compare the developmental potential, 226 were used to compare the pronuclear features (including 95 blastocysts and 131 arrested embryos), 86 1PN embryos were processed the genetic testing (including 69 blastocysts and 17 arrested embryos) in this study. The flow chart of screening subjects was shown in Figure 1.

Figure 1. The flow chart of screening subjects.
Ovarian stimulation, oocyte retrieval, insemination, fertilization check, and embryo evaluation
The protocols of ovarian stimulation, oocyte retrieval, and insemination were performed as previously described with a small modification (Xiong et al., Reference Xiong, Han and Liu2011; Ye et al., Reference Ye, Huang and Zeng2009).
For the IVF insemination: Each cumulus-oocyte complex was inseminated in a 50 ul equilibrated IVF microdroplet at 39∼41 h post-HCG, using 10,000∼15,000 motile sperm per microdroplet. Cumulus cells were mechanically removed after 5-h short co-incubation of gametes. Zygotes with two distinguished PBs were defined as fertilization and were transferred to the fresh cleavage medium (G1, Vitrolife Sweden AB, Sweden) in the time-lapse incubators (EmbryoScope Plus, Vitrolife, Sweden) for further incubation. Oocytes with only one polar body were left in an IVF dish containing sperm overnight. In the cycles with lower fertilization rate (< 30%) or total fertilization failure, oocytes without the second polar body received rescue ICSI at 5–6 h post insemination.
For the ICSI insemination: Cumulus cells were removed by exposure to 60 IU/ml of hyaluronidase for 30–60 s. ICSI was performed at 39∼41 h post-HCG by three experienced embryologists. Oocytes were immediately transferred to the time-lapse incubators after ICSI.
Fertilization was checked 18–20 h after insemination. 1PN zygotes were cultured in the time-lapse incubators until day 5 or 6. Three-pronuclear zygotes were not further cultured. From day 1 to day 3, embryos were cultured in a G1 medium. From day 3 to day 5 or 6, embryos were cultured in a blastocyst medium (G2, Vitrolife Sweden AB, Sweden). The embryo quality was evaluated on day 3, day 5, and day 6 based on the previous criteria (Gardner & Lane, Reference Gardner and Lane1997).
The arrested embryos means those which were at cleavage-stage and did not reach blastocyst stage until day 6.
Time-lapse live embryo imaging and the observation of pronuclear characteristics
All embryos were monitored in the time-lapse incubators at our clinic since January 2020, beginning 5 h after IVF, and immediately after ICSI. The time-lapse images were captured automatically at 10-min intervals until the blastocyst stage.
The images of zygotes were carefully replayed to rule out ‘false’ 1PN on the fertilization checking day. The area and diameter of the pronuclear and the number of NPB were recorded when they reached the maximum. We measured the area and diameter using the software provided by the time-lapse incubator system.
Blastocyst biopsy, vitrification, and pre-implantation genetic testing procedures
Blastocysts with a grade of AA, AB, BA, or BB were defined as good quality, with a grade of BC, CB, or CC were defined as poor quality. The biopsy protocol was performed as previously described (Xiong et al., Reference Xiong, Liu and Wang2021). Briefly, on day 5 or 6, a ∼ 10-μm hole was drilled on the ZP immediately before biopsy with the laser. The holding pipette aspirated the blastocysts with the ICM at the 7 o’clock position. An opening was made on the ZP at the 3 o’clock position. After the blastocyst collapsed and the TE cells crumbled, the 4-6 TE cells were aspirated into the biopsy pipette. Finally, the TE cells were removed by a quick flicking movement of the biopsy pipette against the holding pipette. The TE cells were thrice-rinsed in G-mops medium and then transferred to the Polymerase Chain Reaction (PCR) tubes for further processing. Blastocysts were vitrified immediately after biopsy and TE cells were processed for genetic testing.
In the first stage from December 2021 to February 2022, the next-generation sequencing (NGS) technique was used to test the biopsied cells. In the second stage from March 2022 to September 2022, the single nucleotide polymorphisms (SNP) method was used to test the biopsied cells in order to identify the uniparental diploidy.
After WGA with the WGA kit (genome sequencing universal sample processing kit, Yikon Genomics, Suzhou, China), DNA samples were subjected to NGS on the Illumina Hiseq 2500 platform (Illumina, San Diego, CA, USA) with 2.0-Mb raw reads generated for each sample. ChromGo (Yikon Genomics) software was used to analyze sequencing data and report chromosomal abnormalities. Biopsy diagnostic decisions were based on copy number variations: a chromosome with two copies was assigned a value of 2 and determined to be diploid, a chromosome with one copy was assigned a value of 1 and determined to be a haploid, and a chromosome with three copies was assigned a value of 3 and determined to be a triploid.
Statistical analyses
Statistical analysis was performed using Stata (version 12.0). Quantitative data was presented as the mean ± standard deviation (SD). Independent t-test was used for the comparison between two groups, analysis of variance was used for the comparison among multiple groups. P value < 0.05 was considered statistically significant.
Results
The overall 1PN rates were 3.41% (1250/36691) during the study period: 3.46% (975/28184) for IVF cycles, and 3.23% (275/8507) for ICSI cycles, respectively (P = 0.715). Only 1PN zygotes exhibiting intact and clear pronuclear images were included in this study.
The characteristics of cycles
The characteristics of cycles with or without 1PN were compared. The cycles with 1PN zygotes showed significantly different demographic characteristics to those without 1PN zygotes, in terms of couples’ age, attempts, causes of infertility, stimulation protocols, oocytes yield, and transferrable embryos on day 3 (Table 1).
Table 1. The characteristics of cycles with or without 1PN zygotes

The embryonic development of 1PN zygotes
A total of 388 1PN zygotes were utilized to assess developmental potential. The overall blastocyst formation and good-quality blastocyst rates were 22.94% (89/388) and 16.24% (63/388), respectively, which were significantly lower than their cohort 2PN controls (63.25% (10305/16292) and 50.23% (8184/16292), respectively, P = 0.000).
Accordingly, the corresponding rates in IVF-derived 1PN zygotes were 27.69% (67/242) and 19.83% (48/242), significantly higher than that of ICSI-derived 1PN zygotes (8.90% (13/146) and 6.16% (9/146), respectively, P < 0.005). These data indicated that 1PN zygotes had compromised developmental potential, especially for those originating from ICSI cycles.
The pronuclear characteristics of 1PN zygotes
In this section, a total of 226 1PN zygotes, comprising 95 blastocysts and 131 arrested embryos, and 151 cohort 2PN zygotes, including 85 blastocysts and 66 arrested embryos, were enrolled to evaluate pronuclear characteristics, including pronuclear area and diameter, as well as the number of nucleolar precursor bodies (NPB) (Table 2).
Table 2. The pronuclear characteristics of 1PN and 2PN zygotes
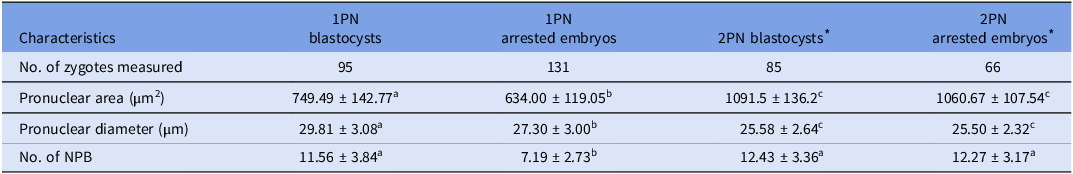
Note: Values with superscript letters a, b and c were statistically different across groups (P < 0.05).
* The diameter represented one of the pronuclear in 2PN zygotes; the pronuclear area included both of the two pronuclear; the number of NPB included both of the NPB in two pronuclear.
The findings revealed that 1PN blastocysts showed significantly larger pronuclear area (749.49 ± 142.77 vs. 634.00 ± 119.05, P = 0.000), extended pronuclear diameter (29.81 ± 3.08 vs. 27.30 ± 3.00, P = 0.000), and an increased number of NPB (11.56 ± 3.84 vs. 7.19 ± 2.73, P = 0.000), in contrast to 1PN arrested embryos. However, 2PN blastocysts and arrested embryos displayed similar pronuclear characteristics. The pronuclear area of 1PN zygotes was significantly smaller than that of 2PN zygotes (the aggregate pronuclear area in two pronuclei). It is noteworthy that the NPB count in 1PN blastocysts was comparable to that of their cohort 2PN blastocysts (11.56 ± 3.84 vs. 12.43 ± 3.36, P = 0.055); conversely, the NPB count in 1PN arrested embryos was significantly reduced compared to 2PN arrested embryos (7.19 ± 2.73 vs. 12.27 ± 3.17, P = 0.000).
These data demonstrated that 1PN zygotes that developed into blastocysts had significantly different pronuclear characteristics compared to arrested embryos; their NPB count were comparable to the cohort 2PN controls although the pronuclear area were smaller.
The genetic analysis of 1PN embryos
In this section, eighty-six 1PN embryos underwent genetic analysis, including 69 blastocysts and 17 arrested embryos; the remaining 17 blastocysts were excluded due to financial constraints.
Among the 1PN blastocysts, 95.65% (66/69) were diploidy, 4.35% (3/69) were haploidy. The 1PN blastocysts displayed significantly elevated diploidy euploidy rates (66.67 vs. 11.76% (2/17), P = 0.000), markedly reduced aneuploidy rates (23.19 vs. 88.24% (2/15), P = 0.000), in comparison to 1PN arrested embryos. Four cases of uniparental diploid were identified by SNP. Interestingly, all originated from ICSI cycles.
The genetic outcomes were further analyzed by different insemination and testing methods. Regarding different insemination methods, 75.44% of 1PN blastocysts in IVF cycles were diploidy euploidy and had reproductive potential, which were significantly higher than those from ICSI cycles (75.44 vs. 25.00%, P = 0.001). For different testing methods, 35 blastocysts were tested by NGS method. The results exhibited that 65.71% (23/35) was diploidy euploidy, 31.43% (11/35) was aneuploidy, and 2.86% (1/35) was haploidy. The remaining 34 blastocysts were tested by SNP. Of which, 67.65% (23/34) was diploidy euploidy, 14.71% (5/34) was aneuploidy, 5.88% (2/34) was haploidy, and 11.76% (4/34) was uniparental diploidy. The diploidy euploidy rates, aneuploidy rates and haploidy rates were comparable between the two testing methods (65.71 vs. 67.65%, 31.43 vs. 14.71%, and 2.86 vs. 5.88%, respectively, P > 0.05).
Additionally, the genetic results in 1PN blastocysts were compared to those of 2PN blastocysts (from PGT-A cycles with females aged <35 years). The results showed that the diploidy euploidy rates in 1PN blastocysts were comparable to those of 2PN blastocysts (66.67 vs. 59.74%, P = 0.260, Table 3).
Table 3. The chromosome constitution of embryos from 1PN zygotes
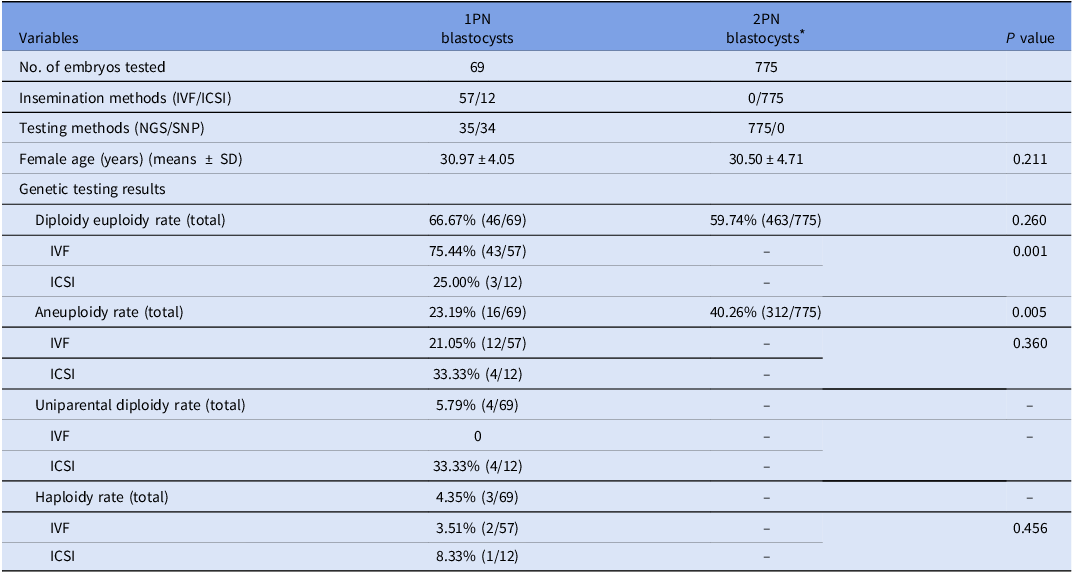
* The 2PN blastocysts were selected from PGT-A cycles with females aged <35 years during the study period.
The correlation between pronuclear characteristics and chromosome constitution in 1PN blastocysts
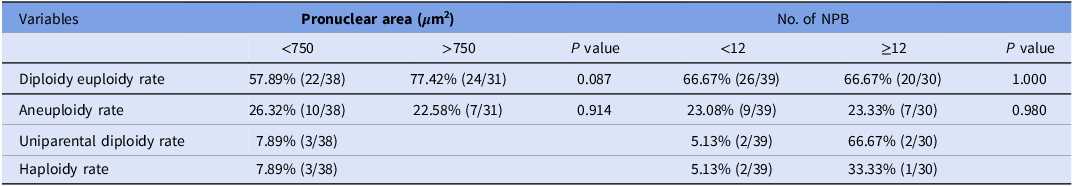
The correlation between pronuclear characteristics and chromosome constitution in 1PN blastocysts were investigated (Table 4). Regarding pronuclear area, the 1PN blastocysts with a larger pronuclear area (>750 μm2) tended to exhibit a higher diploidy euploidy rate compared to those with a smaller area (<750 μm2), yet this disparity did not reach statistical significance (77.42 vs. 57.89%, P = 0.087). For the NPB count, the 1PN blastocysts with a greater number of NPB (≥12) showed equivalent diploidy euploidy rates compared to those with fewer NPB (< 12) (66.67 vs. 66.67%, P = 1.000).
Table 4. The correlation between pronuclear area, number of nucleolar precursor body (NPB) and chromosome constitution in 1PN blastocysts
Discussion
The present study demonstrated that the developmental potential of 1PN zygotes was compromised, which was correlated to their pronuclear characteristics. Those 1PN zygotes developed to blastocysts stage exhibited distinctive pronuclear characteristics, including larger area, longer diameter, and elevated NPB count, compared to arrested embryos. The 1PN embryos displayed high diploidy rates. The diploidy euploidy rates in 1PN blastocysts were comparable to those of 2PN blastocysts, and were significantly higher in IVF-derived 1PN blastocysts compared to ICSI-derived 1PN blastocysts. However, no significant association was identified between pronuclear characteristics and their chromosome composition.
The characteristics of cycles, including female age, stimulation protocols, E2 on hCG trigger day, oocytes yield, number of oocytes matured were significantly different between cycles with or without 1PN zygotes. These results were in line with previous report (Fabozzi et al., Reference Fabozzi, Rega and Starita2019).
It is noteworthy that 15 1PN zygotes were reclassified as 2PN when the time-lapse images were retrospectively analyzed, indicating the asynchrony appearance or disappearance of pronuclei. It has been reported that up to 25% of 1PN zygotes in IVF should be re-designated as 2PN following an observation 4-6 h after the standard fertilization check (Staessen et al., Reference Staessen, Janssenswillen and Devroey1993). However, majority of studies checked pronuclear at a single time point due to the absence of time-lapse incubators (Fu et al., Reference Fu, Chu and Zhou2022; Li et al., Reference Li, Dang and Wang2020; Xie et al., Reference Xie, Tang and Hu2018; Hirata et al., Reference Hirata, Goto and Izumi2020; Kai et al., Reference Kai, Moriwaki and Yumoto2018). The dynamic review of the time-lapse images avoided the inclusion of ‘false’ 1PN zygotes and enhanced the precision of our findings.
The embryonic development of 1PN zygotes was firstly evaluated. Generally, 1PN zygotes exhibited markedly reduced developmental potential compared to 2PN controls, particularly for those originating from ICSI cycles. These results are in accordance with previous findings (Araki et al., Reference Araki, Itoi and Honnma2018; Fu et al., Reference Fu, Chu and Zhou2022; Itoi et al., Reference Itoi, Asano and Shimizu2015).
It is well known that the pronuclear morphometry, including area and diameter, is associated with the developmental potential in 1PN zygotes (Araki et al., Reference Araki, Itoi and Honnma2018), with 1PN blastocysts presented with larger pronuclear and diameter. A comparable result was found in the present study.
In addition to the diameter and area of pronuclear, we questioned whether the quantity of nucleolar precursor body (NPB) was correlated with the developmental potential of 1PN zygotes. Our data confirmed this hypothesis. The findings indicated that the NPB count in 1PN blastocysts was significantly higher compared to that of the 1PN arrested embryos, and was on par with the cohort 2PN controls.
It has been reported that the 1PN might derive from the fusion of the sperm and the female counterpart when the entry of the sperm takes place within 18 μm from the second polar body (Wei et al., Reference Wei, Enatsu and Furuhashi2022). It is not difficult to understand that when sperm entry is in close proximity to the spindle, it might lead to the fuse of female and male pronuclei when male chromosomes bump into their female counterparts due to the cytoplasmic wave (Wei et al., Reference Wei, Enatsu and Furuhashi2022). These pronuclei presented an enlarged area and an increased NPB count because they encompassed both female and male genomes. Conversely, arrested 1PN embryos had a reduced pronuclear area and a diminished NPB count, suggesting impaired pronuclear assembly and abnormal mitotic division.
The present study showed that the haploidy rate was only 4.00% and the diploidy rate reached 90.00% in 1PN embryos. These results are alignment with previous studies, where the diploidy rate ranges from 62 to 87% (Hirata et al., Reference Hirata, Goto and Izumi2020; van der Heijden et al., Reference van der Heijden, van den Berg and Baart2009; Sultan et al., Reference Sultan, Munné and Palermo1995). The different aetiologies of formation could account for the divergent chromosomal ploidy (Wei et al., Reference Wei, Enatsu and Furuhashi2022), including (1) Parthenogenetic activation of the oocyte; (2) Oocyte activation with sperm entry, followed by female pronuclear formation, with male pronuclear failing to form; (3) Male pronuclear formation, with female pronuclear failing to form; (4) Female and male pronuclear formation that is fused, creating one enlarged pronuclear encapsulating both female and male genomes; and (5) Single pronuclear membrane formation encapsulating both female and male genomes. The first three items should be haploid, and the latter two should be diploid (Wei et al., Reference Wei, Enatsu and Furuhashi2022). The elevated diploidy rate observed in 1PN embryos indicates that the majority of 1PN zygotes may originate from the incorporation of female and male genomes.
Furthermore, the diploidy euploidy rates in 1PN blastocysts were comparable to those of their 2PN controls, significantly higher than that of arrested embryos. This result was in line with previous studies (Xie et al., Reference Xie, Tang and Hu2018; Hirata et al., Reference Hirata, Goto and Izumi2020; Soler et al., Reference Soler, Bautista-Llàcer and Escrich2021). It is easy to understand that 1PN embryos, originating from abnormal fertilization or via parthenogenic activation, are more prone to arrest before blastulation; whereas those with normal chromosome complements are more likely to progress to the blastocyst stage.
Previous studies have demonstrated that IVF-derived 1PN blastocysts exhibit significantly higher euploidy rates than ICSI-derived 1PN blastocysts (Staessen et al., Reference Staessen, Janssenswillen and Devroey1993; Mateo et al., Reference Mateo, Vidal and Parriego2017; Mateo et al., Reference Mateo, Parriego and Boada2013). A comparable result was obtained in our study. The formation mechanism might account for their different chromosomal constitution. In IVF cycles, when the fusion of the sperm takes place within 18 μm from the second polar body, male chromosomes might bump into their female counterparts due to the cytoplasmic wave to form 1PN (Wei et al., Reference Wei, Enatsu and Furuhashi2022). Those 1PN embryos contain both female and male genomes, and presented with diploidy euploidy chromosome (Krukowska & Tarkowski, Reference Krukowska and Tarkowski2005). ICSI-1PN, however, mainly derives from the close of the sperm injection to the spindle, which might result in an error in pronuclear formation, abnormal mitosis, and thus chromosomal abnormalities (Mateo et al., Reference Mateo, Parriego and Boada2013).
Forty-six 1PN blastocysts had no detectable chromosomal abnormalities in this study. Of these, 23 blastocysts were confirmed to have a biparental chromosomal constitution by SNP analysis, which were suitable for clinical use. The remaining 23 blastocysts were subjected to sequencing by NGS, further testing was required to ascertain biparental genetic contribution prior to clinical use. These findings suggest that 1PN blastocysts possess the clinical utility following genetic testing, particularly for those patients without other 2PN embryos available.
It is noteworthy that all the uniparental diploidy (n = 4) were exclusively identified in ICSI-derived 1PN blastocysts, with no such cases detected in IVF-derived 1PN blastocysts. Whether it is sufficient to subject only ICSI-derived blastocysts to SNP analysis while selecting IVF-derived blastocysts based on their pronuclear characteristics, warrants further investigation.
We aimed to establish whether the pronuclear features are associated with chromosome ploidy in the 1PN blastocysts. However, no significant relationship was identified between pronuclear area, NPB count, and chromosome constitution. It has been reported that t8 (time to eight cells), t9 (time to eight cells), and tEB (time from insemination to expanded blastocyst) are valuable morphokinetic indicators for predicting ploidy in 2PN embryos (Bamford et al., Reference Bamford, Barrie and Montgomery2022). Further research is necessary to explore the applicability of these indicators for predicting the ploidy of 1PN embryos.
This study, however, has certain limitations. The correlation between pronuclear characteristics and genetic constitution was investigated only in a limited number of embryos. The modest sample size may have attenuated the strength of our conclusions. Moreover, some 1PN blastocysts were analyzed by NGS, which has its own constraints as it may not detect uniparental diploidy. Further prospective studies with larger cohorts are necessary to verify our findings.
Funding standards
This work was supported by the Chongqing Science and Technology Committee (grant number: CSTB2022NSCQ-MSX0253), Chongqing Health Committee (grant number: 2021MSXM108), Yuzhong District Science and Technology Committee (grant number: 20190143), and Women and Children’s Hospital of Chongqing Medical University (grant number: 2021YJQN07).
Competing interests
The authors declare that they have no conflict of interest.
Ethical standards
The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration.
Ethics approval
This study was approved by the Institutional Review Board (IRB) and the Ethics Committee of Women and Children’s Hospital of Chongqing Medical University (Ethics approval number: 2021-RGI-10). Every patient in this study provided written informed consent.
Consent for publication
Written informed consent for publication was obtained from all participants.








