Schizophrenia is a severe mental illness affecting approximately 1% of the population worldwide.Reference van Os and Kapur1 Previous linkage and twins studies suggest a strong genetic component to schizophrenia, with its heritability being estimated at 80%.Reference McGuffin, Gottesman, Swerdlow, Binder and Parker2 However, the culprit genes and the biological mechanism involved in the pathogenesis of schizophrenia remain unclear.Reference Van Os, Rutten and Poulton3 Recently, the Psychiatric Genomics Consortium performed a genome-wide association study (GWAS) in a sample of 38 131 patients with schizophrenia and 114 674 controls (known as PGC-1); 108 genes, including both novel genes and those previously reported, were identified.Reference Ripke, Neale, Corvin, Walters, Farh and Holmans4 However, these common susceptibility loci passing the genome-wide significance level only explain approximately 3.4% of the heritability of schizophrenia,Reference Ripke, Neale, Corvin, Walters, Farh and Holmans4 and the replication from other GWAS is only modest. Although some studies have provided evidence that joint contributions from several common variants, each with modest effects, explain a small fraction of the heritability for complex diseases,Reference Visscher, Brown, McCarthy and Yang5 it is possible that many common variants related to schizophrenia have not achieved a genome-wide significance in the GWAS because of small effect size or insufficient sample size. Indeed, according to the hypothesis that the interaction between various genes underlies the majority of complex diseases,Reference Wilkinson, Evgrafov, Zheng, Hartel, Knowles and Graham6 even those variants with a small effect size may provide pivotal information on the biological basis of the disease.
The neuroimaging of schizophrenia
The inconsistent replications among many GWAS of schizophrenia so far might be because of the limited statistical power arising from the small sample size and the clinical heterogeneity among recruited patients with schizophrenia. One feasible strategy to overcome these barriers is to use endophenotypes or intermediate phenotypes in molecular genetic studies. Endophenotypes are considered to be more proximal to the biological aetiology of a disorder,Reference Gottesman and Gould7 and might provide an alternative strategy in the identification of the pathogenesis of schizophrenia.Reference Wang, Xiang, Deng, Wu, Li and Ma8 For instance, previous magnetic resonance imaging (MRI) studies reported significant abnormalities in the white matter integrity of patients with schizophrenia when compared with healthy controls, and this finding has been supported by a meta-analysis study.Reference Dong, Wang, Chang, Jiang, Klugah-Brown and Luo9 Furthermore, fractional anisotropy, a measure reflecting the fibre bundle connectivity of white matter, has been found to be significantly low in patients with schizophrenia compared with healthy controls,Reference Wang, Cheung, Deng, Li, Huang and Ma10 especially in brain regions such as the genu of the corpus callosum, posterior cingulum fibres, superior and inferior fronto-occipital fasciculus and the posterior corona radiata.Reference Kunimatsu, Aoki, Kunimatsu, Abe, Yamada and Masutani11 In fact, some other studies have indicated that fractional anisotropy is a highly heritable trait and could be used as a potential quantitative endophenotype in understanding the aetiology of schizophrenia.Reference Skudlarski, Schretlen, Thaker, Stevens, Keshavan and Sweeney12
In the present study, instead of targeting the individual gene, we hypothesised that the abnormal endophenotypes of schizophrenia might be mainly associated with the dysfunctional gene pathways or networks,Reference Wilkinson, Evgrafov, Zheng, Hartel, Knowles and Graham6, Reference Schijven, Kofink, Tragante, Verkerke, Pulit and Kahn13 the identification of which could provide pivotal information on the biological mechanism for schizophrenia.
Building on our hypothesis, we aim to (a) examine fibre bundle connectivity of white matter indexed by fractional anisotropy values in schizophrenia and (b) explore the association between rare variants in gene pathways or networks and abnormal white matter integrity in schizophrenia.
In the current study, we first performed voxel-wise comparisons in a sample set of first-episode and treatment-naïve patients with schizophrenia and matched controls by diffusion tensor imaging (DTI), and identified the brain regions with abnormal fractional anisotropy values. Each differential fractional anisotropy value and a composite index of overall fractional anisotropy value computed by principal component analysis (PCA) were then used as one of endophenotypes in the subsequent genetic analysis. Furthermore, by leveraging the summary statistics generated from the GWAS of schizophrenia in PGC-1,Reference Ripke, Sanders, Kendler, Levinson, Sklar and Holmans14 we carried out a gene-wise subnetwork analysis to identify the pathways/networks associated with schizophrenia, followed by a weighted gene correlation network analysis,Reference Langfelder and Horvath15 finding modules in 15 brain regions that are highly correlated with genes in the associated pathway/network. Finally, in each identified module, association analysis was carried out to identify the genes associated with the value of each differential and overall fractional anisotropy value in an attempt to explore the potential role of genetic variants in schizophrenia, specifically, its fibre bundle disconnectivity.
Method
Participants
A total of 240 participants comprising 100 first-episode, treatment-naïve patients with schizophrenia and 140 healthy controls were recruited at the Mental Health Centre of the West China Hospital, Sichuan University, China. The Structured Clinical Interview for DSM-IV, Patient VersionReference First, Spitzer, Gibbon and Williams16 was used by a trained psychiatrist to interview and diagnose patients with schizophrenia. Patients initially diagnosed with the schizophreniform disorder were followed for at least 6 months to confirm the diagnosis of schizophrenia. All patients were evaluated with the Positive and Negative Syndrome Scale for their disease severity.Reference Kay, Flszbein and Opfer17 Healthy controls were recruited from the local area by poster advertisements and were screened for a lifetime absence of psychiatric illnesses with the Structured Clinical Interview for DSM-IV, Non-Patient Version.Reference First, Spitzer, Gibbon and Williams16 All participants were Han Chinese from the Sichuan province of China. The study was approved by the Ethics Committee of the West China Hospital of Sichuan University 2017 (approval number 131). All participants provided written informed consent for participation in the study.
MRI data acquisition, preprocessing and imaging processing
Whole-brain diffusion-weighted images were recorded along 15 gradient directions (b = 1000 s/mm2, number of excitations: 2) together with one unweighted (b = 0) image (42 images in total). Each image was acquired using a single-shot spin echo-planar imaging sequence (repetition time: 10 000 ms, echo time: 70.8 ms, slice thickness: 3.0 mm with no gap, field of view: 240 mm2, matrix size: 256 × 256, voxel resolution: 0.94 × 0.94 × 3 mm3). Raw data were qualitatively inspected by an experienced neuroradiologist. No gross abnormalities were observed in any participant.
High-resolution T1 images were obtained by three-dimensional spoiled gradient echo sequence from all participants. The sets used in this protocol included the following: repetition time: 8.5 ms; echo time: 3.93 ms; dip angle: 12u; thickness of slice: 1 mm, single shot; field of view: 24 cm × 24 cm; matrix: 256 × 256; size of vowel: 0.47 × 0.47 × 1 mm3. A total of 156 axial image slices were collected per brain.
DTI data were preprocessed by the FMRIB's Diffusion Toolbox within FMRIB software library (FSL) (http://www.fmrib.ox.ac.uk/fsl). Image preprocessing included the following steps: (a) the effects of head motion and image distortion caused by eddy currents were corrected by applying an affine alignment to register all other diffusion images to the b0 images in the original DTI data; (b) the non-brain tissue and background noise were removed from the b0 image by using FSL brain extraction tool (BET) command on the eddy corrected image; (c) the diffusion tensor for each voxel was estimated by the multivariate linear fitting algorithm, and the tensor matrix was diagonalized to obtain three pairs of eigenvalues (L1, L2 and L3) and eigenvectors; (d) the voxel-wise value of fractional anisotropy was calculated for each participant, and (e) fractional anisotropy maps of all participants were normalised to a 2 × 2 × 2 mm3 Montreal Neurological Institute standard space, and smoothed with a 6 mm full-width at half-maximum Gaussian kernel.
Total intracranial volume was estimated with FreeSurfer software (http://surfer.nmr.mgh.harvard.edu/fswiki), which includes a set of automated tools, to reconstruct the intracranial volume from the T1-weighted MRIs.Reference Buckner, Head, Parker, Fotenos, Marcus and Morris18 More detailed information was described in our previous study.Reference Xiang, Wu, Wang, Li, Jiang and Deng19
The voxel-wise comparisons of fractional anisotropy values were performed with Statistical Parametric Mapping (version SPM8; https://www.fil.ion.ucl.ac.uk/spm/) processing, and two sample t-tests were performed to explore the main effects with gender, age and total intracranial volume as covariates. Statistical inferences were made with a voxel-level threshold of P < 0.05 after false discovery rate correction for multiple comparisons.
Genome-wide genotyping and quality control
DNA was extracted from the whole-blood samples of 100 patients with schizophrenia and 140 healthy controls through a standard phenol-chloroform isolation method. The HumanOmniZhongHua-8 Bead Chip was chosen to genotype these blood samples. The generated genotyping data with a high calling rate were subsequently filtered according to genotype rates, minor allele frequency (MAF) and Hardy–Weinberg equilibrium tests (in controls only). Single nucleotide polymorphisms (SNPs) with a missing rate per individual >5%, with an MAF <0.05 and SNPs failing the Hardy–Weinberg equilibrium tests (P-value ≤ 10–5 (please superscript)) were excluded from the study (148 480 SNPs).
Consequently, a total of 742 805 SNPs with a mean call rate of 98.9% passed the quality control. Seven patients and six controls were excluded from the study because of a missing rate per SNP of >3%, or the inconsistency between the chromosomal X status and self-reported gender information, or because of the cryptic relatedness test (proportion identity-by-descent > 0.2). Finally, 93 patients diagnosed with schizophrenia and 134 controls were included in the subsequent analyses. EIGENSTRAT 3.0Reference Price, Patterson, Plenge, Weinblatt, Shadick and Reich20 software was used to detect population stratification, which applies PCA to capture hidden population structures in GWAS data (Supplementary Figure 1 available at https://doi.org/10.1192/bjp.2018.297). The top three principal components were extracted as covariates in subsequent association analyses.
Whole-exome sequencing and variants calling
A total of 234 out of 240 participants (97 patients and 137 controls) were sequenced with the TruSeqExome Enrichment Kit, optimised for Illumina HiSeq2000 sequencing. Three patients and three controls were unable to be sequenced because of poor DNA quantity. Data preprocessing included (a) the Burrows–Wheeler Alignment toolReference Li and Durbin21 to reference against the human genome (hg19); (b) Picard tools (http://picard.sourceforge.net/) to collect quality statistics and fix read group issues; (c) Samtools (http://samtools.sourceforge.net/) to filter out low-quality reads, and GATKReference McKenna, Hanna, Banks, Sivachenko, Cibulskis and Kernytsky22 for indel alignment, and SNP and indel calling. Quality control of all variants and samples is presented in the Supplementary Material.
Identifying significant gene-wise subnetworks associated with schizophrenia
The summary statistics from PGC-1, generated from a GWAS of 9394 patients with schizophrenia and 12 462 controls,Reference Ripke, Sanders, Kendler, Levinson, Sklar and Holmans14 were downloaded (https://www.med.unc.edu/pgc) and used to identify significant gene-wise subnetworks associated with schizophrenia. A gene-based analysis of the summary statistics was conducted, using the VEGAS program with default parameters,Reference Liu, Mcrae, Nyholt, Medland, Wray and Brown23 the output of which was then integrated with a human interactome network,Reference Menche, Sharma, Kitsak, Ghiassian, Vidal and Loscalzo24 using protein interaction network-based pathway analysisReference Wang, Matsushita, Madireddy, Mousavi and Baranzini25 to identify the enriched subnetworks. The statistical threshold was set at <0.1 (fewer than 10% of random networks result in a significant subnetwork) after permutation testing (shuffling the data 1000 times).Reference Wang, Matsushita, Madireddy, Mousavi and Baranzini25
Constructing gene co-expression networks in brain regions
The gene co-expression networks in brain regions were constructed with the whole-genome transcriptomic data from BrainSpan (http://www.brainspan.org/); the data was collected by RNA-sequencing, and the gene expression was defined by a normalised reads per kilobase million value of 1 in at least one region at one time point for 80% of the available samples. In the current study, the expression data of 15 brain regions, including 11 neocortical regions, the striatum, the hippocampus, the thalamus and the amygdaloid, were chosen to construct the gene co-expression networks, using the R package ‘WGCNA’Reference Langfelder and Horvath15 (Supplementary Table 1).
Gene ontology enrichment analysis
WebGestaltReference Wang, Duncan, Shi and Zhang26 was used to perform gene ontology enrichment analysis of genes in each module. A hypergeometric test implemented in WebGestalt computed the enrichment P-value, followed by a Benjamini–Hochberg correction for multiple testing. Enriched gene ontology terms are reported at a Benjamini–Hochberg-corrected P < 0.05.
Cell-specific expression analysis
To detect the cell type overrepresented by the genes from each network and by taking advantage of an online tool (CSEA) for cell-specific expression analysis (http://genetics.wustl.edu/jdlab/csea-tool-2/),Reference Dougherty, Schmidt, Nakajima and Heintz27 we conducted a cell-specific enrichment analysis for the genes in each network module. In brief, a large survey of central nervous system cell-specific microarray data was used to identify those genes that are significantly enriched in each population data and provide a simple perusable archive of plots of this measure across all cell types. The algorithms are available online (www.bactrap.org).Reference Dougherty, Schmidt, Nakajima and Heintz27
Association analysis of fractional anisotropy and overall fractional anisotropy
We performed PCA with oblimin rotation for differential fractional anisotropy values, which showed a significant difference between patients with schizophrenia and controls, and the first component was extracted as overall fractional anisotropy. Taking into account the fact that these differential fractional anisotropy values are correlated with each other and with overall fractional anisotropy, the correlation analyses among the five original fractional anisotropy variables, and between the overall fractional anisotropy and each differential fractional anisotropy variable were conducted. The SNP-set (Sequence) Kernel Association Test (SKAT) program,Reference Wu, Kraft, Epstein, Taylor, Chanock and Hunter28 which accounts for both linear and nonlinear interactions, was used to test the association of common and rare variants of genes in each module with the value of each differential fractional anisotropy and the overall fractional anisotropy, respectively, with age, gender, years of education and the top three principal components served as covariates. In an attempt to control the type I error in the identification of the rare variants in the genes constituting each module associated with each differential fractional anisotropy and the overall fractional anisotropy, a permutation test was used to obtain empirical P-values. In our analysis, the rare variants were defined as those with an MAF <1% from the whole-exome sequencing data, and the common variants as those with an MAF ≥5% from the GWAS data. Genetic loci (single nucleotide variations) mapped to genes were extracted based on whole-exome sequencing and GWAS data in each module. Phenotypes and genetic loci were then randomly shuffled, leaving the correlation of phenotype with genotype unchanged. This process was repeated 10 000 times and obtained 10 000 results (Zn).Reference Wu, Kraft, Epstein, Taylor, Chanock and Hunter28 The empirical P-value for the association of genes in each module with the overall fractional anisotropy was evaluated by calculating the proportion of the Zn less than or equal to the overall observed Zns.
Results
Demographic data and clinical characteristics
The demographic and clinical characteristics of all participants who completed the clinical assessments and the whole-brain diffusion-weighted images in the study are shown in Table 1. There appeared no significant differences in mean age (t = −0.285, P = 0.776), gender distribution (Pearson's χ2 = 0.011, P = 0.918) and years of education (t = −0.831, P = 0.407) between patients with schizophrenia and healthy controls.
Table 1 Demographic and clinical characteristics of patients with schizophrenia and healthy controls
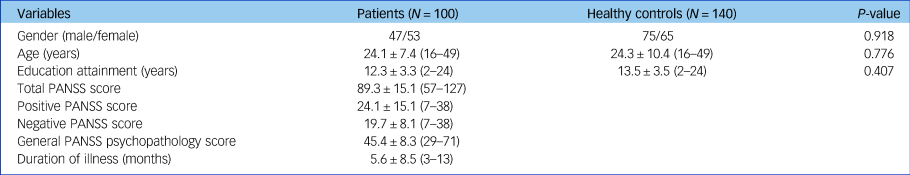
Data are displayed as mean ± s.d. (range), unless otherwise indicated. PANSS, Positive and Negative Syndrome Scale.
Voxel-wise comparisons of fractional anisotropy
Fractional anisotropy was significantly reduced in the left and right anterior cingulate cortex, the left and right precuneus and the extra-nuclear in patients with schizophrenia compared with controls (false discovery rate-corrected P < 0.01, cluster size ≥100; Supplementary Figure 2, Table 2). The overall fractional anisotropy was obtained by extracting the first component of the PCA from the fractional anisotropy values of the above five regions. There was a significant correlation among the five original fractional anisotropy variables (Supplementary Table 2), and between the overall fractional anisotropy and all of the five original fractional anisotropy variables (Supplementary Table 3).
Table 2 Fractional anisotropy differences between patients with schizophrenia and controls
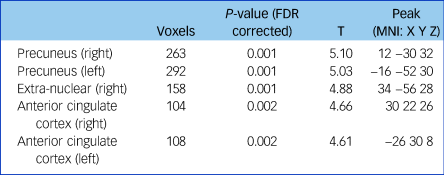
Threshold at P < 0.01 (FDR corrected) and cluster size ≥100. FDR, false discovery rate; MNI, Montreal Neurological Institute.
Gene-wise subnetworks and co-expressed gene modules
A subnetwork including 207 genes was identified by the network-based analysis of the summary statistics from PGC-1 (Supplementary Figure 3 and Table 4). The co-expression network analysis found that 176 out of the 207 genes passed the quality control and were highly co-expressed in four gene modules (named yellow, blue, brown and turquoise; Fig. 1, Supplementary Table 5). Supplementary Table 6 displays the gene ontology enrichment of genes in each module. The yellow module contained co-expressed innate immune response genes (10 out of 24, 41.6%), major histocompatibility complex (MHC) class I receptor activity genes (4 out of 24, 16.6%) and MHC class I protein complex genes (5 out of 24, 20.8%). It is noteworthy that the average expression of genes in the yellow module is increased mainly during the early prenatal development (post-conception weeks 13–37; Supplementary Figure 4). The blue and brown modules predominantly contain genes functioning in histone methyltransferase activity and histone-lysine N-methyltransferase activity. The turquoise module contains genes related to phosphotransferase activity, alcohol group as acceptor and DNA binding. In addition, genes in the yellow module indicated a significantly increased cell type-specific enrichment in astrocytes (P = 0.002, P adjusted = 0.04).
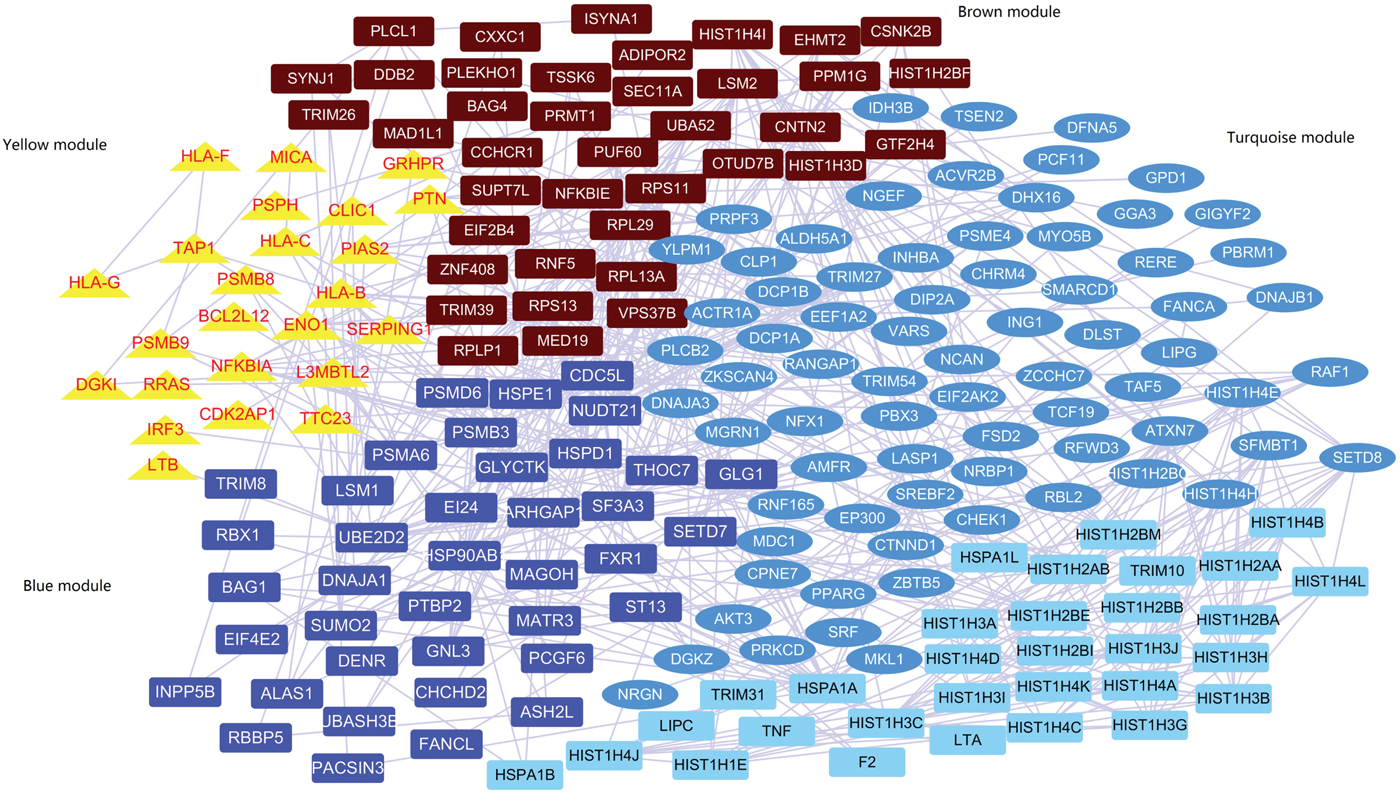
Fig. 1 Co-expression module containing genes in protein–protein interaction.
Gene-set association test in each module
The cumulative evidence of rare or common variants for all genes in each module was evaluated in patients with schizophrenia and controls. Using the whole-exome sequencing data, we found that the combining effect rare variants of genes in the yellow module were significantly associated with schizophrenia (P = 0.002, P adjusted = 0.008). Furthermore, we found that the same combining effect in the yellow module was also associated with reduced fractional anisotropy in the left anterior cingulate cortex (P = 0.006, P adjusted = 0.024) and with reduced overall fractional anisotropy (P = 0.041, uncorrected) only in patients with schizophrenia, not in the controls (Table 3). However, no significant association between common variants of genes in each module and the overall fractional anisotropy was detected.
Table 3 Cumulative rare variants of genes in each module were associated with the reduced left anterior cingulate cortex
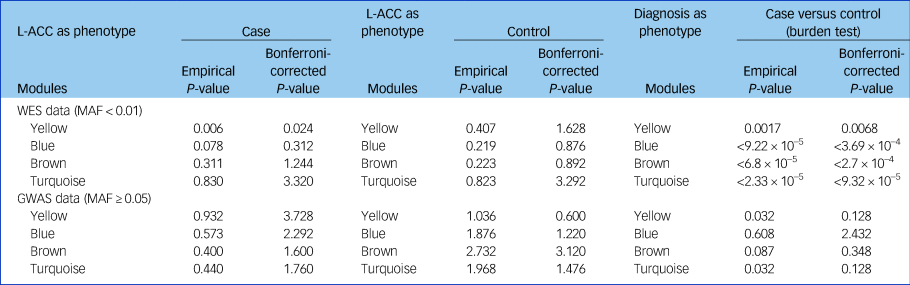
GWAS, genome-wide association study; L-ACC, left anterior cingulate cortex; MAF, minor allele frequency; WES, whole-exome sequencing.
Discussion
In the present study, we found that the first-episode, treatment-naïve patients with schizophrenia had significantly reduced fractional anisotropy in the left and right anterior cingulate cortex, the left and right precuneus and the extra-nuclear compared with healthy controls. By integrating the results of the PGC-1 GWAS for schizophrenia, interactome data and brain co-expression networks, a subnetwork with 207 genes were found to be associated with schizophrenia; of these, 176 genes were highly co-expressed in four gene modules. Furthermore, we found that the rare variants in the genes constituting the yellow module, mainly the genes related to the immune pathways, were associated with reduced fractional anisotropy in the anterior cingulate cortex only in patients with schizophrenia. Our findings thus further support the hypothesis that gene pathways or networks might substantially contribute to the abnormal endophenotypes in schizophrenia.
The abnormal fractional anisotropy of white matter in patients with schizophrenia identified in this study reflects a disruption of neural circuitry.Reference Kunimatsu, Aoki, Kunimatsu, Abe, Yamada and Masutani29 Our findings are consistent with previous studies, including our own,Reference Lei, Li, Deng, Li, Huang and Ma30 suggesting that the impaired fibre bundle connectivity between the brain regions plays a critical role in the neuropathology of schizophrenia. Further identification of genes or pathways involved in neural connectivity circuits could deepen our understanding of the complex causes of schizophrenia.Reference Gupta, Chen, Liu, Damaraju, Wright and Perrone-Bizzozero31
The same approach has been taken previously to identify the genetic subnetworks associated with other complex diseases; for example, Han et al integrated the GWAS and human protein interaction networks to identify a subnetwork of 39 genes that was not only enriched for genes associated with alcohol dependence, but also collectively associated with alcohol dependence in three independent samples.Reference Han, Yang, Kranzler, Liu, Zhao and Farrer32 In the current study, following the identification of a subnetwork with 207 genes associated with schizophrenia, we found that 176 of those genes were highly co-expressed in four gene modules. Each module represents particular gene ontology biological processes, molecular functions and cellular components. Of note, the yellow module was found in our data to be associated with reduced fractional anisotropy in the left anterior cingulate cortex, and it is also enriched by genes related to MHC class I (HLA-B, HLA-C, HLA-F and HLA-G) and genes (DGK1, IRF3, MICA, PIAS2 and TAP1) previously reported to be associated with schizophrenia.Reference Andrews and Fernandez-Enright33, Reference Li, Zhang, Lencz, Darvasi, Alkelai and Lerer34
It is worth highlighting here that innate immune response genes, such as MHC class I receptor activity genes and MHC class I protein complex genes, were significantly enriched in the yellow module. MHC class I belongs to the MHC gene family,Reference Shiina, Hosomichi, Inoko and Kulski35 which is encoded by three genes in humans: HLA-A, HLA-B and HLA-C. It has previously been reported that MHC class I molecules were expressed in neurons in the central nervous system throughout developmental period and into adulthood, and may mediate the contributions of genetic and environmental factors to schizophrenia through direct yet temporal-specific effects on neurite outgrowth, synapse formation and function, long-term and homeostatic plasticity, and activity-dependent synaptic refinement.Reference McAllister36 Other than MHC class I, some other genes in the yellow module are also worth notice. The interferon regulatory factor 3 gene (IRF3), located on chromosome 19q13, plays a key role in the innate immune system through the response to viral infection.Reference Collins, Noyce and Mossman37 Melnik et al reported that IRF3 might be critical in regulating the development of neuronal progenitor cells.Reference Melnik, Tauber, Dumrese, Ullrich and Wolf38 The analysis of protein–protein interaction networks showed that IRF3 interacts with other schizophrenia susceptibility genes, such as CREB1, AKT1 and ESR1.Reference Li, Zhang, Lencz, Darvasi, Alkelai and Lerer39
Furthermore, several studies identified that AKT1 and CREB1 were associated with synaptic plasticity in the hippocampus.Reference Levenga, Wong, Milstead, Keller, LaPlante and Hoeffer40 Liu et al identified that PIAS1 has an important role in neuronal plasticity, learning and memory function.Reference Liu, Ma and Lee41 Also, CDK2AP1 in the yellow module was recently identified as a susceptibility gene for intellectual disability.Reference Bulayeva, Lesch, Bulayev, Walsh, Glatt and Gurgenova42 Alsayegh et al have also shown that CDK2AP1 gene was associated with the differentiation of human embryonic stem cells and the percentage of cells in the S phase.Reference Alsayegh and Sheridan43 On the other hand, Manolio et al reported that the cumulative effect of rare variants, especially genes regulating immune activation, may influence brain structure and neurodevelopment deficit in patients with schizophrenia as well as account for the part of ‘missing heritability’ in schizophrenia.Reference Manolio, Collins, Cox, Goldstein, Hindorff and Hunter44 A recent next-generation sequencing study, including 4877 patients with schizophrenia and 45 376 controls from Sweden, confirmed the effects of rare variant burden in patients and contributed to the understanding of the genetic architecture and biological patterns of schizophrenia.Reference Genovese, Fromer, Stahl, Ruderfer, Chambert and Landén45
As shown in this study, the expression of genes in the yellow module increased during early prenatal development, with a significant cell type-specific enrichment in astrocytes. The present study, in line with previous studies, provides another piece of evidence that brain development and immune dysregulation in the prenatal environment affect the pathology of schizophrenia in a temporal- and cell-specific interaction.Reference Brown46 Further, animal studies also provide supportive evidence. For example, the previous study by Bauman et al reported that prenatal exposure to maternal immune activation might alter brain structure and behaviour related to schizophrenia in offspring in rhesus monkeys.Reference Bauman, Iosif, Smith, Bregere, Amaral and Patterson47 Another study showed that prenatal exposure to maternal inflammation influences the levels of N-acetylaspartate/creatine and myo-inositol/creatinine in the cingulate cortex of mice.Reference Li, Leung, Zhou, Ho, Kong and Basil48 In fact, abnormal white matter microstructure in the left anterior cingulate cortex has been found in mice exposed to an immune challenge in early or late prenatal development,Reference Li, Cheung, Wei, Cheung, Hui and You49 which is consistent with our findings in the current study.
Using data from the PGC-I, we found that the blue and brown modules harboured the genes predominantly related to the histone methyltransferase activity and the histone-lysine N-methyltransferase activity. It has previously been reported that multiple immune system-related genes and genes related to histone H3-K4 methylation are associated with schizophrenia and bipolar disorder.50 In agreement with previous studies, the current findings provide further evidence for the role of histone methylation in schizophrenia. Furthermore, previous studies have shown that the histone methylation pathway may coordinate complex cognitive processes involved in long-term memory in schizophrenia, depression, autism and neurodegenerative disease,Reference Jarome and Lubin51 and could play an important role in the interaction between environmental factors and genetics on susceptibility to psychiatric manifestations throughout life.Reference Ibi and González-Maeso52
By studying first-episode and treatment-naïve patients with schizophrenia, we minimised the effects of confounding factors such as chronicity of the illness and antipsychotic treatment. The main limitation of this study is the moderate sample size, which limited statistical power, especially for common variants with small or medium effects. Further replication in a larger and independent sample is required.
In summary, through an integrated analysis of genetic data from the PGC-I for schizophrenia, interactome data and brain co-expression networks, the current results indicate that dysregulation of genes involved in the immune system play an important role in the pathogenesis of schizophrenia. The combining effect of the rare variants in the genes making up the yellow module, especially genes related to the immune system, was associated with abnormal fractional anisotropy of white matter in patients with schizophrenia. These findings provide some important clues as to the aetiology of schizophrenia. Moreover, the present study also highlights the feasibility of using endophenotypes in exploring the pathogenesis of neuropsychiatric diseases such as schizophrenia.
Supplementary material
Supplementary material is available online at https://doi.org/10.1192/bjp.2018.297.
Funding
This work was partly funded by National Key Research and Development Program of the Ministry of Science and Technology of China grant 2016YFC0904300 (to T.L.); National Nature Science Foundation of China Key Project (81630030 and 81461168029 to T.L.); National Nature Science Foundation of China Project (81771446 to Q.W.; 81701322 to W.L.); 1.3.5 Project for disciplines of excellence, West China Hospital of Sichuan University (ZY2016103 and ZY2016203 to T.L.); and the optimal individualised treatment options for antipsychotics from the Ministry of Science and Technology of China grant (2016YFC1307005 and 2016YFC1307000 to Q.W.).







eLetters
No eLetters have been published for this article.