Introduction
Proteases play crucial roles in the life cycle of various helminthic parasites. Parasite proteases are likely to be participated in host–parasite interactions and parasite metabolism, for instance, host tissue invasion and evasion of host immune response (Na et al. Reference Na, Kim, Lee, Kim, Bae, Kang, Yu, Sohn, Cho and Kong2006). Serine protease inhibitors (serpins) are members of a superfamily proteins identified in a broad range of living beings, including viruses, bacteria, animals and plants (Irving et al. Reference Irving, Pike, Lesk and Whisstock2000; Gettins, Reference Gettins2002), with a characteristic well-conserved tertiary structure (Roberts et al. Reference Roberts, Mathialagan, Duffy and Smith1995). Serpins participate in numerous fundamental biological functions such as blood coagulation (Debrock and Declerck, Reference Debrock and Declerck1997; Sugino et al. Reference Sugino, Imamura, Mulenga, Nakajima, Tsuda, Ohashi and Onuma2003), signalling cascades (O'Donnell and Blackman, Reference O'Donnell and Blackman2005), fibrinolysis (Mak et al. Reference Mak, Enghild and Dubin1996), inflammation (Thorgersen et al. Reference Thorgersen, Ghebremariam, Thurman, Fung, Nielsen, Holers, Kotwal and Mollnes2007), activation of the complement system (Congote, Reference Congote2007) and in mechanisms associated with host immune modulation (Chopin et al. Reference Chopin, Matias, Stefano and Salzet1988). Numerous serpins have been identified in parasitic helminths that were involved in immune regulation and parasite survival via interference with the host immune-stimulatory signals (Molehin et al. Reference Molehin, Gobert and McManus2012).
T aenia solium is an important zoonotic parasite that causes a great threat to public health and economic loss in developing countries (Fleury et al. Reference Fleury, Carrillo-Mezo, Flisser, Sciutto and Corona2011). Despite quite a few researches regarding the biology and immunology of T. solium, the mechanisms associated with host immune evasion or the roles of serpins in these processes were still unknown. In other organisms, serpins contributed to inhibiting blood coagulation, damaging host proteases, and escaping from host immune attacks. In parasitic helminths, serpins appeared to play the similar roles (Molehin et al. Reference Molehin, Gobert and McManus2012). Additionally, serpins were of considerable value as a diagnostic tool. Schistosoma mansoni serpins, Sm-SERPIN and Sm-RP26, showed significantly high reactivity to sera from S. mansoni-infected patients and were diagnostic of that particular species (Tanigawa et al. Reference Tanigawa, Fujii, Miura, Nzou, Mwangi, Nagi, Hamano, Njenga, Mbanefo, Hirayama, Mwau and Kaneko2015). The Schistosoma haematobium serpin was also considered as a marked species-specific antigen (Blanton et al. Reference Blanton, Licate and Aman1994). Serpins also had been shown to be highly immunogenic, displaying strong reactivity to sera from infected rats (Molehin et al. Reference Molehin, Gobert, Driguez and McManus2014), suggesting that the immunodiagnostic potential of the serpins stemmed from the important physiological roles in host–parasite interactions. In view of the immunodiagnostic potential of serpins, the possible diagnostic value of Tsserpins should be explored.
Through the analysis of previously published T. solium genome and transcriptome data (Tsai et al. Reference Tsai, Zarowiecki, Holroyd, Garciarrubio, Sanchez-Flores, Brooks, Tracey, Bobes, Fragoso, Sciutto, Aslett, Beasley, Bennett, Cai, Camicia, Clark, Cucher, De Silva, Day, Deplazes, Estrada, Fernández, Holland, Hou, Hu, Huckvale, Hung, Kamenetzky, Keane, Kiss, Koziol, Lambert, Liu, Luo, Luo, Macchiaroli, Nichol, Paps, Parkinson, Pouchkina-Stantcheva, Riddiford, Rosenzvit, Salinas, Wasmuth, Zamanian, Zheng, Cai, Soberón, Olson, Laclette, Brehm and Berriman2013) and structure-to-function bioinformatics analysis of the Tsserpins’ polypeptides, five Tsserpins were produced as recombinant proteins in Escherichia coli system and identified as diagnostic candidates. The indirect enzyme-linked immunosorbent assays (iELISAs) were established based on the five recombinant proteins to explore a sensitive and specific assay for the diagnosis of porcine cysticercosis.
Materials and methods
Ethics statement
This study did not involve the use of endangered or protected species, and was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences. All experiments were conducted maintaining current China laws. The protocol was approved by the Committee on the Ethics of Animal Experiments of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences.
Parasite materials and serum samples
Healthy pigs of 40–60 days aged without cysticercosis were purchased from a local area in Gaolan, Gansu Province, China, and all pig blood samples were collected as negative control by saphenous bleeding before infection. Thirty pigs were orally infected with 3000 eggs of T. solium that was collected from Dali, Yunnan Province, China. Blood samples were collected by saphenous bleeding 60 days post-infection (dpi) (pigs were confirmed by necropsy examination). Trichinella spiralis-positive sera were from six pigs orally infected with 2000 infective muscle larvae (ML) at 35 dpi (pigs were confirmed by necropsy examination). The sera against Toxoplasma gondii were obtained from six pigs that were inoculated peritoneally with 1000 oocysts. T. gondii infection was confirmed by polymerase chain reaction (PCR) as described previously (He et al. Reference He, Ma, Song, Zhou, Wang, Huang and Zhu2016) at 15 dpi.
Cysticercus tenuicollis positive sera (six samples) were collected from naturally infected pigs from local slaughterhouse in Yongdeng, Gansu Province, China. Pigs were provided with non-medicated feed and water ad libitum in the experiment, housed in ventilated pigsty and monitored every day. At the end of experiments, all pigs were humanely sacrificed, and sera samples were collected and stored at −20 °C until use. Meanwhile, the parasites were isolated and stored at −80 °C until use in other experiments.
Identification of Tsserpins
Source sequences encoding Tsserpins were analysed utilizing BLAST (Basic Local Alignment and Search Tool) with the BLASTP and BLASTN algorithm against the Taenia Genome Database (TGD) available at http://taenia.big.ac.cn/taenia/index.html and T.solium gene annotation (http://192.168.51.199/index.html). The serpins sequences were confirmed by inspecting the expected length range of 350–450 amino acids (Molehin et al. Reference Molehin, Gobert and McManus2012) and the presence of two highly conserved motifs (NAVYFKG and DVNEEG) (Han et al. Reference Han, Zhang, Min, Kemler and Hashimoto2000).
In silico analysis
The Tsserpins coding sequences were compared with known entries in GenBank using the BLASTP program. The physicochemical property of the mature Tsserpins including theoretical molecular weight, isoelectric points and amino acid (aa) composition were calculated using the ProtParam software (http://web.expasy.org/protparam/). To gain insight on probable functionality, the deduced T.solium serpins amino acid sequences were scanned against amino acid motif entries, PROSITE, ScanProsite, SignalP and TMHMM servers (ExPASY bioinformatics Resource Server; http://www.expasy.org/proteomics). Putative N-glycosylation sites were identified by NetNGly1.0 server (http://www.cbs.dtu.dk/services/NetNGlyc/) and phosphorylation sites were predicted by NetPhos2.0 server (http://www.cbs.dtu.dk/services/NetPhos/). The Hydrophilicity Plot-Kyte-Doolittle, Flexible Regions-Karplus-Schulz, Antigenic Indes-Jameson-Wolf, Surface Probability-Emini were analysed employing DNAStar 7.1 software (DNAStar, USA) to predict the potential linear B-cell epitopes of T.solium serpins.
Structure modelling
The secondary structure of T.solium serpins were predicted using PredictProtein (http://www.predictprotein.org/). The three-dimensional (3D) of Tsserpins and other serpins were predicted by Swiss-Model program. QMENA was used to estimate model reliability and predict quality. The predicted structures were aligned and viewed using DeepView-SWISS pdbViewer v4.1.
Phylogenetic analysis
Amino acid sequences of Tsserpins and the representative parasite serpins from GenBank (accession numbers in Fig. 2) were submitted to the ClustalW 1.83. The phylogenetic analyses were performed by MEGA6.0 for tree building and TreeView for tree drawing.
Total RNA extraction and cDNA synthesis
Total RNA was extracted from 30 mg T.solium cysticerci using Trizol reagent (Invitrogen, USA), according to the manufacturer protocol. All reagents for RNA extraction were prepared using diethypyrocarbonate (DEPC) treated water. The concentration and integrity of extracted RNA samples were determined with a Nandrop® spectrophotometer (Thermo Scientific). cDNA was synthesized from total RNA by One-step Reverse Transcription Kit (clontech) following the manufacturer's instruction.
Cloning and expression of Tsserpins
The full-length coding sequences were amplified from T.solium cysticerci cDNA by PCR using specific primers to each of the following Tsserpins: TsB6, TsEP45, Ts570, Ts4848 and Ts12383. Primers were designed using Primer Premier 5 (Table 1S). Individual reactions for PCR amplification contained a premix cocktail (dNTPs, PCR buffer, DNA polymerases) 25 µL, 1·0 µL cDNA, 1·0 µL of each primer (10 µm), 22 µL ddH2O (deionized water). The amplification reactions were carried out using a thermal cycling profile of 95 °C for 5 min; 30 cycles at 94 °C for 40 s, 65 °C for 40 s, 72 °C for 90 s and a final extension for 10 min at 72 °C. The PCR products were analysed on a 1% (w/v) agarose gel and purified by Agrose Gel DNA Extraction Kit (Clontech).
The fragments of Tsserpins were digested by restriction enzymes (Table 1S) and ligated into the pET-30a (+) vector, the plasmids were transformed into E. coli DH5α competent cells and identified by PCR, the positive clones were confirmed by sequencing. Positive recombinant plasmids, termed pET-30a (+)-TsB6, pET-30a (+)-Ts570, pET-30a (+)-TsEP45, pET-30a (+)-Ts4848, pET-30a (+)-Ts12383, were transformed into E.coli BL21 competent cells and cultured to express the recombinant proteins. Lysates from collected bacteria were analysed by sodium dodecyl sulphate (SDS)–polyacrylamide gel electrophoresis (SDS–PAGE).
Protein purification and immunoblotting
Recombinant proteins were purified by a Ni2+ Sepharose™ 6 Fast Flow purification kit as the manufacturer's instructions (GE, USA). Concentrations of purified protein samples were detected by Pierce™ BCA (bicinchonininc acid) protein assay kit (Thermo Scientific, USA) and identified by Western blotting. Briefly, the recombinant proteins were isolated by SDS–PAGE and transferred to nitrocellulose membrane, and then the membrane was incubated with the positive serum from porcine cysticercosis (1:200 dilution) and subsequently incubated with horseradish peroxidase (HRP)-conjugated rabbit anti-pig IgG (1:10 000 dilution, Sigma), the reactions were detected with DAB (diaminobenzidine).
Development of iELISA based on Tsserpins
Purified Tsserpin were diluted in coating buffer (0·05 M carbonate-bicarbonate buffer, pH 9·6) to a final concentration of 5 µg mL−1. The microtitre plates were coated with Tsserpin protein (100 µL well−1) and blocked with 1% (w/v) bovine serum albumin in PBST (Phosphate Buffered Saline with Tween-20) for 1 h at 37 °C. After three times of washing with PBST, the plates were incubated with diluted positive serum samples (1:200) from pigs infected with T. solium (n = 30), C. tenuicollis (n = 6), T. gondii (n = 6), T. spiralis (n = 6) and 51 negative control sera samples, respectively. The plates were washed three times and incubated with HRP-conjugated anti-pig IgG (1:10,000) for 30 min at 37 °C. After washing, the enzyme reaction was detected with TMB (3,3′,5,5′-tetramethylbenzidine) (Sigma, USA). The optical density (OD) value was measured at a wavelength of 450 nm with spectrophotometer (Bio-RAD).
Statistical analysis
Receiver operating characteristics (ROC) (Greiner et al. Reference Greiner, Pfeiffer and Smith2000) analysis was performed to assess the diagnostic value of antigens. One-way ANOVA tests were performed to compare all pairs of groups. Statistical significance was set at P < 0·05. Data analysis was showed using GraphPad Prism 5.0 (GraphPad Software Inc., San Diego, USA).
Results
Cloning and general characteristics of Tsserpins
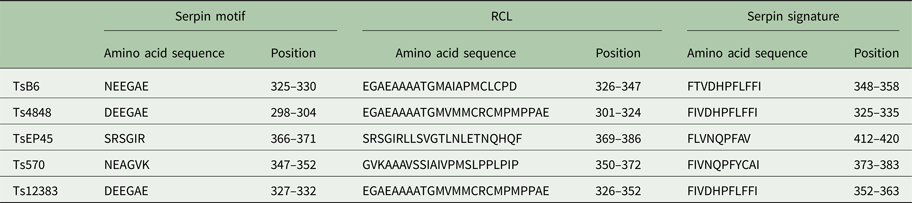
The cloned full-length cDNA and general characteristics of Tsserpins are listed in Table 1. According to the MEROPS (http://merops.sanger.ac.uk/inhibitors/index.shtml) classification of protease inhibitors (Rawlings,et al. Reference Rawlings, Waller, Barrett and Bateman2014), Tsserpins are members of inhibitor family I4 (Clan ID). Multiple sequence alignment of Tsserpin amino acid sequences showed low overall identity (21–69%) with representative serpins from GenBank (Fig. 1). Analysis of Tsserpin peptides indicated the presence of three conserved motifs, the serpin signature, serpin motif and the reactive centre loop (RCL) (Table 2). The cleavage sites of the RCL are located near the C-terminal at position 342 M-343C of TsB6, 344R-345C of Ts12383, 316R-317C of Ts4848, 364 M-365S of Ts570 and 383N-384Q of TsEP45, respectively. Analysis of Tsserpin RCLs (Table 2S) showed that the hinge region contained the typical consensus sequences, with a characteristic ‘P17[E] – P16[E/K/R] – P15[G] – P14[T/S] – P13[X] – P12–9[AGSX] – P8–1[X] – P1′–4′’ common to all inhibitory serpins (Gettins, Reference Gettins2002),where the amino acid residues are defined as …P3–P2–P1(cleavage site)–P1′ – P2′ – P3′…and X is any amino acid. The RCL of TsB6, Ts4848 and Ts12383 were highly similar to typical pattern of RCL, while Ts570 was with residue variation only at position P16 and P14. Unexpectedly, TsEP45 differed greatly from the patterns of other serpins, which showed several variations: P14 [I] and P12−9 [LLSV]. No signal peptide or transmembrane domain was found in the peptide sequences. Other post-translational modification sites, such as cAMP- and cGMP-dependent protein kinase phosphorylation, casein kinase II phosphorylation, N-myristoylation, protein kinase C phosphorylation and tyrosine kinase phosphorylation sites were predicted to present in the five Tsserpins (Table 3S). According to the analysis of hydrophilicity, flexible regions, antigenic index and surface probability, potential B cell epitopes also might be present in all Tsserpins (Table 4S).

Fig. 1. Alignment of partial Tsserpin amino acid sequences with those of other representative serpins. Sh (Schistosoma haematobium), Em (Echinococcus multilocularis), Eg (Echinococcus granulosus), Sj (Schistosoma japonicum), Sm (Schistosoma mansoni), Cs (Clonorchis sinensis), Pw (Paragonimus westermani), Bm-1 and Bm-2 (Brugia malayi), Hc (Haemonchus contorus), Tv (Trichostrongylus vitrinus), A-psin(Antitrypsin), T-mb (Thrombin), T-psin (chyMotrypsin), As (Ascaris suum), Tg (Toxoplasma gondii), Tsp (Trichinella spiralis), TsB6, Ts570, TsEP45, Ts4848, Ts12383 (Taenia solium). All GenBank accession numbers are shown in Fig. 2. The conserved residues were in red underlines; the residues of reactive center loop are bordered in black boxes.
Table 1. Predicted general characteristics of Tsserpins
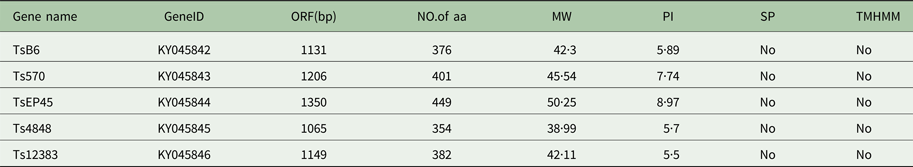
SP, Signal peptide; TMHMM, Transmembrane.
Table 2. Key characteristic serpin amino acid sequences in T. solium
Phylogenetic analysis showed that the Tsserpins were distributed on different evolutionary branches (Fig. 2). Tsserpins all had the high-sequence homology to Echinococcus granulosus serpins: TsB6 shared 67% sequence identity with SerpinB9 (Accession number EUB60253.1), Ts4848 and Ts12383 shared 79% to E.granulosus Serpin (Accession number CDS22753.1), Ts570 shared 75% with E.granulosus Serpin (Accession number EBU63679.1). TsEP45 was closely related to E.granulosus Serpin (Accession number EUB63680·1) with 81% sequence identity. Tsserpins were distantly related to those of other helminths, especially the evolutionary status of TsEP45 and Ts570 were very ancient, far away from the serpins of several common porcine parasites, including Ascaris suum, T. gondii and T. spiralis.

Fig. 2. Phylogenetic tree based on amino acid sequence of serpins from different species by maximum likelihood. The red dots represented T.solium.
Structure-based alignment analysis of Tsserpins
The potential tertiary structure of a protein reveals several specific biological functions. 3D protein structures of Tsserpins were predicted by homology modeling. Firstly, template structures were developed by SWISS-MODEL. The search results (Fig. 3) indicated that TsB6, Ts4848 and Ts12383 shared similar structure to SerpinB3 (2zv6, Homo sapiens, human), the structure of TsEP45 was similar to the serine protease inhibitor (3sto, Schistosoma haematobium), and Ts570 had a similar structure to anithrombin-III (2hij, H. sapiens, human). Ramachandran plotting indicated that the structure models were optimal (data not shown).

Fig. 3. Predicted tertiary structures of Tsserpins. 3A: TsEP45; 3B: Ts570; 3C: Ts4848; 3D: Ts12383; 3E: TsB6; The purple was represented the RCL. The green and red were represented the β sheet, the blue was represented the α helices. The figure shows the Tsserpins in their native conformation with the RCL being surface accessible by target proteases.
Expression and identification of Tsserpins
The PCR products were obtained (Fig. 1S) and inserted into pET-30a (+) vector. Then the recombinant plasmids pET-30a (+)-Tsserpins were confirmed by enzyme digestion (Fig. 2S) and DNA sequencing.
The recombinant pET-30a (+)-Tsserpins were expressed and analysed by Western blotting. SDS–PAGE analysis showed the recombinant proteins were mainly found in the pellet (Fig. 3S). The peak expression was observed at 37 °C when induced with IPTG (isopropyl-β-d-thiogalactoside) at 1·0 mm L−1, following 6 h induction. Tsserpins were purified by Ni2+-affinity chromatography, and a single band for each of recombinant proteins in SDS–PAGE indicated that the recombinant serpins were free of obvious contaminants (Fig. 4). Immunoblotting results showed Tsserpins were reacted with the positive T.solium cysticercus sera (Fig. 5). And no reaction was observed between Tsserpins and the negative control sera from healthy animals.

Fig. 4. Purification of recombinant Tsserpins proteins. M: Protein molecular weight Markers.
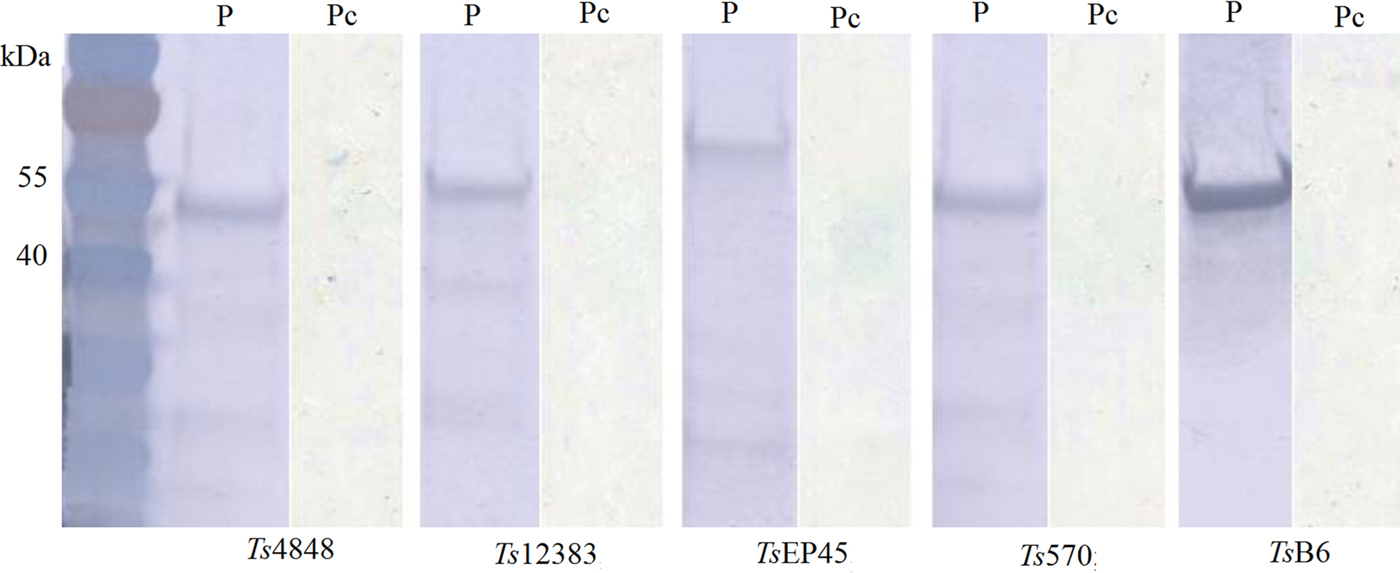
Fig. 5. Immunoblot analysis of recombinant T.solium serpins using the positive serum from porcine Cysticercosis. M: Protein molecular weight Markers, P: the positive serum from porcine Cysticercosis, Pc: Pig negative serum.
Evaluation of the serodiagnostic potential of Tsserpins
To evaluate the possibility of Tsserpin as antigens for diagnosis of cysticercosis, we compared the reactivity of the Tsserpin antigens and various serum samples from pigs experimentally infected Cysticercus cellulosae (n = 30) and negative samples with iELISA. The results showed that TsEP45, TsB6 and Ts570 were specifically reactive with sera from T. solium cysticercosis (Fig. 6). There was no cross-reactivity with positive samples of C. tenuicollis, T. gondii and T. spiralis. The remained two antigens, Ts4848 and Ts12383, detected without disease-specific reactivity (Fig. 6E and 6F), although detective results of positive sera samples from infected pigs with C. cellulosae were higher than those of negative ones.
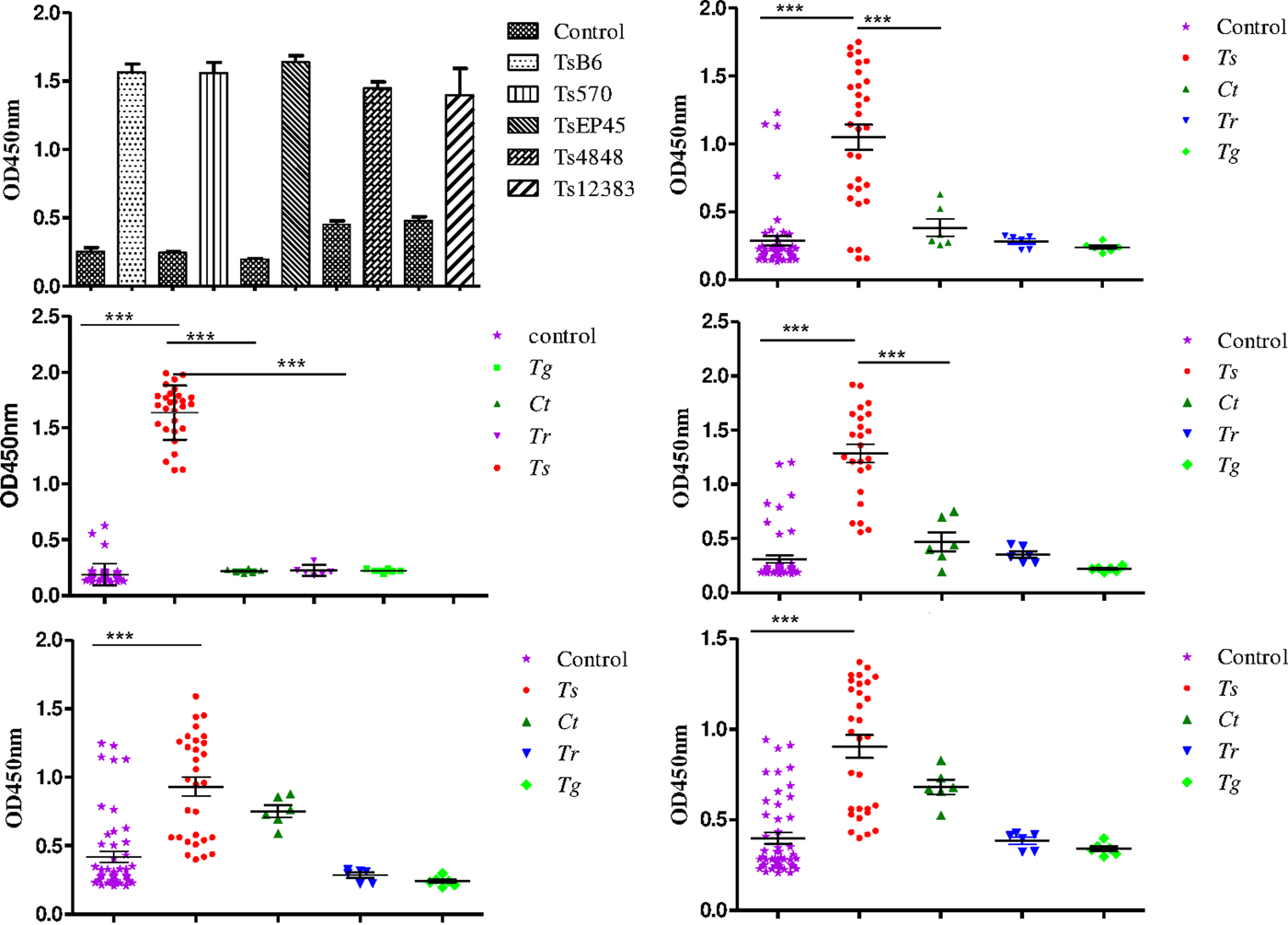
Fig. 6. Species-specific differential detection by Tsserpins. The iELISA was performed using samples from T.solium cysticercus (Ts, n = 30), C. tenuicollis (Ct, n = 6), T. spiralis (Tr, n = 6), and T. gondii (Tg, n = 6) experimentally infected pigs, respectively. Negative control samples (Nc, n = 51) from uninfected areas. The bar lines represent the median while the error bars represent the interquartile range. vertical bars were SEM in panel A. Antigens used in this assay: (6A) the reactivity of the Tsserpins antigens in indirect ELISA; (6B) Ts570; (6C) TsEP45; (6D) TsB6; (6E) Ts4848; (6F) Ts12383. Each data point represents an average of three ELISA replicates. Statistical significance was set at P < 0·05 as depicted using asterisks: * = P < 0·05, ** = P < 0·01, *** = P < 0·001.
To validate of diagnostic performance of iELISAs based on TsEP45, TsB6 and Ts570, ROC analysis was employed for further evaluation. The cut-off values of TsEP45, TsB6 and Ts570 were calculated by sera from 51 uninfected pigs (negative control) and 30 C. cellulosae positive sera. The ROC curve and the corresponding statistics were summarized in Fig. 7. It can be concluded that TsEP45 showed the better diagnostic value than TsB6 and Ts570 and showed acceptable results to be a diagnostic antigen in iELISA.
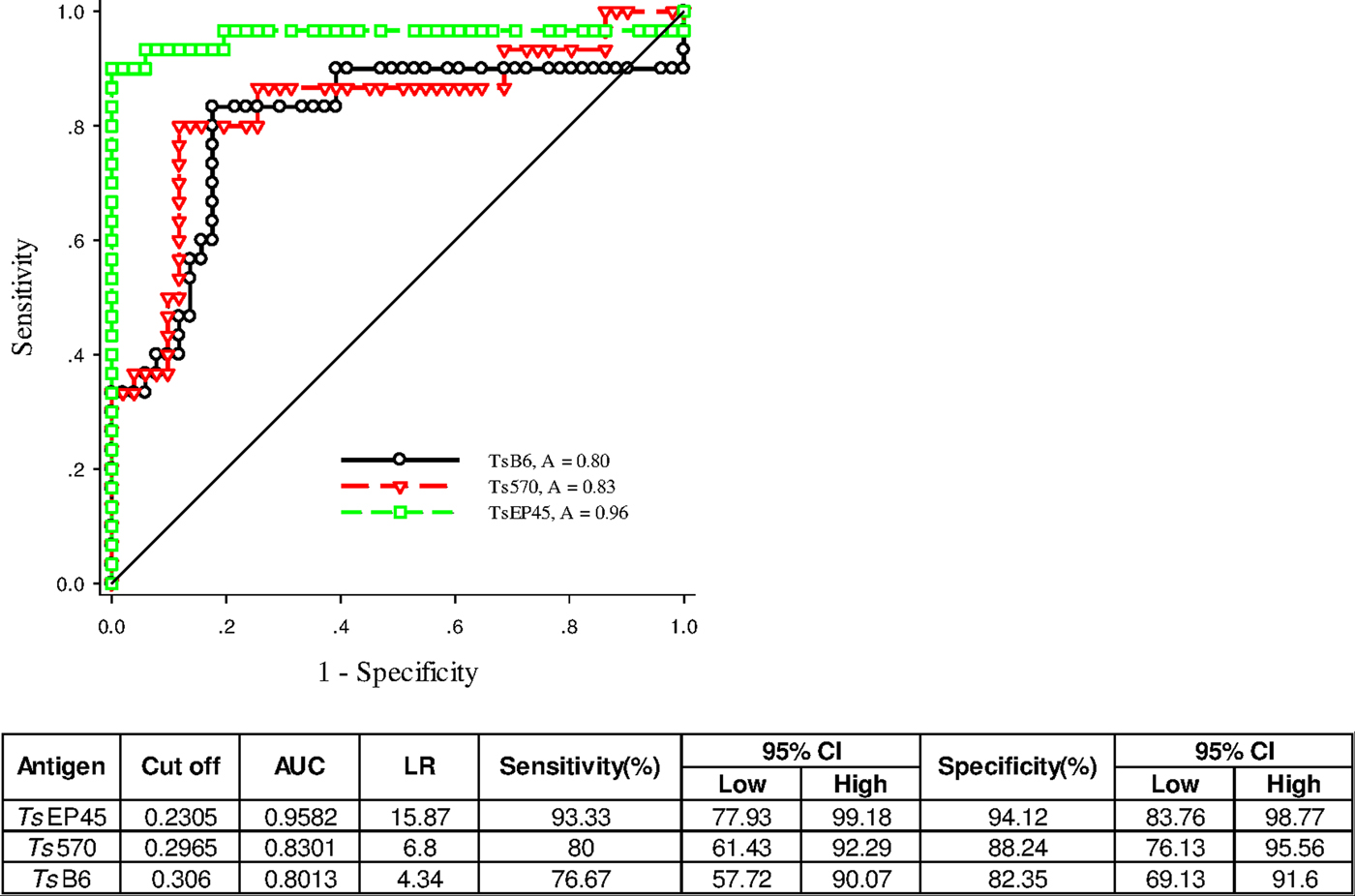
Fig. 7. ROC analysis for TsEP45, TsB6 and Ts570. ROC analysis was performed to determine the area under the curve (AUC), Likelihood ratio (LR), sensitivity and specificity as indicators of the diagnostic value of the TsEP45, TsB6 and Ts570. The TsEP45 showed relatively better diagnostic ability than TsB6 and Ts570. A: AUC, The AUC values close to 1·0 indicate an informative test; and close to 0·5 indicate an uninformative test (Hanley and McNeil, Reference Hanley and McNeil1982); The value of LR greater than 10 (Jaeschke et al. Reference Jaeschke, Guyatt and Sackett1994) practically confirmed the results of diagnosis.
Discussion
Tsserpins were identified as typical serpins due to the presence of characteristic features such as the serpin motif, serpin signature and RCL (Rawlings et al. Reference Rawlings, Waller, Barrett and Bateman2014). Additionally, secondary and tertiary structure prediction analyses indicate that Tsserpins may contain 8–9 α-helices and three β-sheets, features were consistent with the known serpins (Huntington, Reference Huntington2006). The results of the predicted tertiary structure of Tsserpins (Fig. 3) showed that the RCLs of native inhibitory serpins may be exposed and accessible to target proteases, which further strengthens the argument that Tsserpins belong to the serpin superfamily (Rawlings et al. Reference Rawlings, Waller, Barrett and Bateman2014). These observations allowed us to explore Tsserpin biological function based on comparative modelling of known serpins. The conserved 20/21 residue peptide ‘P17[E]–P16[E/K/R]–P15[G]–P14[T/S]–P13[X]–P12−9[AGSX]–P8−1[X]–P1′−4′’ within the RCL determined whether the serpin was inhibitory or non-inhibitory (Irving et al. Reference Irving, Pike, Lesk and Whisstock2000). The primary structures of inhibitory and non-inhibitory serpins have different amino acids of the hinge region (P15 to P8), whereas they are well conserved in inhibitory serpins. In general, P15 is glycine, P14 threonine or serine. P12 to P9, are all alanine residues in most inhibitory serpins. Analysis of Tsserpins showed that TsB6, Ts12383, Ts570 and Ts4848 have typical serpin structures, suggesting that they are functional inhibitory serpins. Further analysis of the hinge regions of TsB6, Ts12383, Ts570 and Ts4848 suggested that they are occupied predominantly by hydrophobic amino acid residues, which are deemed to be the construction of the skeleton conformation needed for the inhibitory activity (McCarthy and Worrall, Reference McCarthy and Worrall1997). However, the corresponding regions of non-inhibitory serpins do not conform to this consensus sequence, for example, TsEP45 shows some variations: P14 [I] and P12–9 [LLSV]. We can speculate, therefore, that TsEP45 may be a non-inhibitory. Non-inhibitory serpins have evolved by ceasing to act as protease inhibitors and by acquiring the ability to interact with other molecules, performing new functions.
To our knowledge, the divergent functions or the specificity of target proteases mainly depends on the variety of RCLs. The target proteases mostly cleave the bonds between P1 and P1′ in RCLs (Silverman et al. Reference Silverman, Bird, Carrell, Church, Coughlin, Gettins, Irving, Lomas, Luke, Moyer, Pemberton, Remold-O'Donnell, Salvesen, Travis and Whisstock2001), which suggested that serpins with different amino acid residues of P1 and P1′ acted on diverse target proteases. According to the reference (Merckelbach and Ruppel, Reference Merckelbach and Ruppel2007) of the amino acid sequence of serpinEmu, R 344 (arginine) can be identified as the P1 residue of Ts12383 and Ts4848, as well as cysteine is found at P1′. Arginine at P1 could explain readily the inhibition of trypsin and pancreatic elastase (PE) since these proteinases cleave their substrates with high specificity after basic amino acids (Merckelbach and Ruppel, Reference Merckelbach and Ruppel2007). Likewise, the intracellular serpin B6 (human) inhibits trypsin via a P1-R (Riewald and Schleef, Reference Riewald and Schleef1996). Therefore, we can envisage that Ts12383 and Ts4848 have potential to inhibit the activity of trypsin and PE. Based on the amino acid sequence of α-1-proteinase inhibitor (α1-antitrypsin), M (methionine) can be identified as the P1 residue of TsB6 and Ts570. Human neutrophil elastase (NE) was inhibited by α1-antitrypsin through P1-M. So, we can speculate that TsB6 and Ts570 have similar function to α1-antitrypsin inhibiting the activity of NE. In summary, P1-P1′ residues were very important in the RCL, and P1 played a crucial role in defining specificity (Potempa et al. Reference Potempa, Korzus and Travis1994). Some serpins were specialized for single proteases, while others inhibited more than one substrate; further in vitro studies could provide evidence of the range of protease inhibition. The predictions above were contributed to a better understanding of T.solium in evading the host immune response. If the Tsserpins were secreted during the oncosphere infection phase, it might be able to block the proteolytic attack of host digestive enzymes, which could be the targets of the intestinal immune system and vaccine candidates.
Phylogenetic analysis showed that Tsserpins are distributed on different evolutionary branches, suggesting the serpins had functional polymorphism and early evolutionary divergence. The Tsserpins in the same branch might share similar function, such as Ts12383, Ts4848 and serpinEmu (Echinococcus multilocularis). In addition, the results of phylogenetic analysis showed that the evolutionary status of TsEP45 was very ancient, far away from the serpins of several common porcine parasites, including A. suum, T. gondii and T. spiralis, suggesting that TsEP45 was a special serpin in common parasites and has potential to act as a species-specific antigen for developing iELISA to detect porcine cysticercosis.
Even though Tsserpins do not contain signal peptide sequences by bioinformatics analysis, they would be released extracellularly through a special pathway independent of the endoplasmic reticulum. Examples from other organisms include human plasminogen-activator inhibitor 2 (Ritchie and Booth, Reference Ritchie and Booth1988) and Hc-serpin from Haemonchus contortus (Yi et al. Reference Yi, Xu, Yan and Li2010). In addition, exosomes were playing crucial roles in cell communication and delivery of components (Valadi et al. Reference Valadi, Ekström, Bossios, Sjöstrand, Lee and Lötvall2007), and it was a possibility that Tsserpins were secreted by exosomes and inducing host immune responses. Based on this initial characterization, Tsserpins had great potential to act as candidate antigens to detect the cysticercus-specific antibodies. To determine the diagnostic potential of Tsserpins in serological assays, the fusion-proteins were coated as antigen in iELISAs. It was found that TsEP45 could effectively differentiate the following parasites species: T. solium, C. tenuicollis, T. gondii and T. spiralis. By detecting sera samples from cysticercus infected pigs, ROC analysis of TsEP45 iELISA exhibited the high sensitivity (93·33%) and specificity (94·12%), respectively. Particularly, the fusion-protein acting as a diagnostic antigen in iELISAs mainly focused on solving the cross-reactivity between C. cellulosae and C. tenuicollis (P < 0·001). The detection results were in accord with sequences alignment results, TsEP45 shared very low similarity to serpins from other species (data not shown). In summary, TsEP45 was a candidate diagnostic antigen for porcine cysticercosis. Further study should be carried out to clarify the relationship between larva loads and dilution of antibody against TsEP45, and how many cysticerci could be detected with this iELISA. The mentioned above are valuable information for establishing a diagnostic method. Meanwhile, iELISA based on TsEP45 will be used to detect the serum samples collected from naturally infected with C. cellulosae and compared with commercial serological methods to demonstrate the feasibility in the future.
Supplementary material
The supplementary material for this article can be found at https://doi.org/10.1017/pao.2017.15
Acknowledgements
The authors would like to thank Professor Baoquan Fu and Professor Delin Zhang (Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences) for offering the serum samples from pigs infected Trichinella spiralis and T. gondii.
Availability of data and material
All sequence data that support the findings of this study have been deposited in GenBank with the following accession numbers: TsB6 (KY045842); Ts570 (KY045843); TsEP45 (KY045844); Ts4848 (KY045845); Ts12383 (KY045846)
Authors’ contributions
GXL and XNL conceived and designed the study, collected and analysed the data, and GXL performed all experiments and wrote the first draft of the paper. PHL and LJW contributed to laboratory work. LM, YDZ, SHZ, AJG, JLH and XNL critically reviewed the manuscript for the important intellectual content. All authors read and approved the final version of the manuscript.
Financial support
This Research was supported by The National Key Research and Development Program of China (grant no. 2017YFD0501303) and the National Natural Science Foundation of China (grant no. 31372433).
Conflict of interest
None.











